The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants - PubMed (original) (raw)
The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants
Katharina Krech et al. Plant Physiol. 2012 Jun.
Abstract
Photosystem biogenesis in the thylakoid membrane is a highly complicated process that requires the coordinated assembly of nucleus-encoded and chloroplast-encoded protein subunits as well as the insertion of hundreds of cofactors, such as chromophores (chlorophylls, carotenoids) and iron-sulfur clusters. The molecular details of the assembly process and the identity and functions of the auxiliary factors involved in it are only poorly understood. In this work, we have characterized the chloroplast genome-encoded ycf4 (for hypothetical chloroplast reading frame no. 4) gene, previously shown to encode a protein involved in photosystem I (PSI) biogenesis in the unicellular green alga Chlamydomonas reinhardtii. Using stable transformation of the chloroplast genome, we have generated ycf4 knockout plants in the higher plant tobacco (Nicotiana tabacum). Although these mutants are severely affected in their photosynthetic performance, they are capable of photoautotrophic growth, demonstrating that, different from Chlamydomonas, the ycf4 gene product is not essential for photosynthesis. We further show that ycf4 knockout plants are specifically deficient in PSI accumulation. Unaltered expression of plastid-encoded PSI genes and biochemical analyses suggest a posttranslational action of the Ycf4 protein in the PSI assembly process. With increasing leaf age, the contents of Ycf4 and Y3IP1, another auxiliary factor involved in PSI assembly, decrease strongly, whereas PSI contents remain constant, suggesting that PSI is highly stable and that its biogenesis is restricted to young leaves.
Figures
Figure 1.
Targeted inactivation of the plastid genome-encoded open reading frame ycf4 by stable transformation of the chloroplast genome. A, Physical maps of the ycf4_-containing region of the tobacco plastid genome and of the transformed plastid genome in Nt-Δ_ycf4 transplastomic lines. All genes shown are transcribed from left to right. Restriction sites relevant for cloning and/or RFLP analysis of transplastomic plants are indicated. Hybridization probes used for Southern-blot and northern-blot analyses are denoted by horizontal bars. The expected sizes of hybridizing fragments in RFLP analysis of wild-type and transplastomic plants are indicated in kb. In Nt-Δ_ycf4_ transplastomic plants, most of the ycf4 coding region is deleted and replaced with the selectable marker gene aadA. The expression of aadA is driven by a chimeric rRNA operon promoter (P_rrn_) and the 3′ untranslated region from the psbA gene (Svab and Maliga, 1993). B, RFLP analysis of transplastomic lines carrying the ycf4 knockout allele. Total cellular DNA was digested with Xho_I and Hin_dIII and hybridized to a radioactively labeled probe detecting the accD region of the plastid genome, which flanks the transgene insertion site. Fragment sizes are given in kb. The size difference of 0.7 kb corresponds to the difference between the wild-type ycf4 gene and the mutant allele disrupted with the aadA selectable marker cassette (compare with A). C, Analysis of ycf4 mRNA accumulation (left panel) and analysis of the expression of petA, an essential gene for Cyt bf function located downstream of ycf4 (right panel), by northern blotting. As a control, RNA from a previously generated knockout line for ycf3, a plastid gene encoding an essential factor for PSI assembly (Ruf et al., 1997), was also loaded. Sizes of the RNA marker bands are given in kb. The absence of ycf4 transcripts from the Nt-Δ_ycf4 transplastomic lines confirms the loss of ycf4 function from the plastid genome. As expected, expression of the aadA selectable marker gene cassette from the strong constitutive P_rrn promoter causes increased accumulation of petA transcripts, due to read-through transcription (Zoubenko et al., 1994; Wurbs et al., 2007). To control for equal loading, the ethidium bromide (EtBr)-stained agarose gels photographed prior to blotting are also shown. WT, Wild type. D, Absence of detectable Ycf4 protein from Nt-Δ_ycf4_ transplastomic plants. To reveal the detection limit of the anti-Ycf4 antibody, a dilution series of thylakoid proteins from wild-type plants was loaded (20 µg chlorophyll = 100%), and the amount of loaded extract from the knockout plant was increased up to 1,000%. To control for loading, a membrane stained with Ponceau red (P.) is shown below the blot.
Figure 2.
Phenotypes of Δ_ycf4_ transplastomic tobacco lines. A, Wild-type (WT) and Δ_ycf4_ transplastomic knockout plants are shown after 8 weeks of growth in soil under low-light conditions (40–50 µE m−2 s−1). For comparison, a PSI-deficient mutant generated by knockdown of the gene encoding the reaction center subunit PsaA (KD-psaA; for details, see text) is also shown. B, After extended growth under low-light conditions (for 16 weeks), Δ_ycf4_ transplastomic lines reached the reproductive stage but developed only very few flowers.
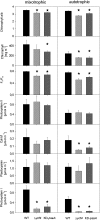
Figure 3.
Analysis of chlorophyll contents and various photosynthetic parameters in transplastomic Δ_ycf4_ knockout mutants and wild-type (WT) as well as KD-psaA control plants. Data sets are shown for plants grown under 40 to 50 μE m−2 s−1 in either synthetic Suc-containing medium (mixotrophic conditions) or soil (autotrophic conditions) for 6 weeks. For each plant line, three or more different plants were measured, and data were subjected to one-way ANOVA using a pairwise multiple comparison procedure (Holm-Sidak method) in SigmaPlot. Highly significant differences are indicated by asterisks (P < 0.05). Error bars represent
sd
. Photosynthetic complexes were quantified from difference absorbance measurements of cytochrome _b_559 (PSII), Cyt bf, plastocyanin, and P700 (PSI) in isolated thylakoids.
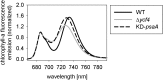
Figure 4.
The 77 K chlorophyll a fluorescence emission measurements. The fluorescence emission signals were normalized to the PSII emission maximum at 685-nm wavelength. The PSI emission signal peaks at 733 nm in the wild type (WT). The blue-shifted wavelength of the PSI emission maximum in the Δ_ycf4_ knockout mutants and the KD-psaA control plants indicates that PSI antenna organization is altered and suggests that a significant proportion of PSI antenna proteins are not attached to PSI core complexes.
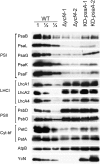
Figure 5.
Immunoblot analysis of diagnostic components of the multiprotein complexes in the thylakoid membrane in transplastomic Δ_ycf4_ knockout mutants, wild-type plants (WT), and KD-psaA control plants grown under autotrophic conditions. Isolated thylakoid proteins were separated by SDS-PAGE, blotted, and probed with antibodies against the proteins indicated on the left side of each panel. Equal amounts of chlorophyll were loaded. For quantitative assessment of protein accumulation in the mutants, a dilution series of the wild-type sample (100%, 50%, and 25%) was loaded. A Ycf4-specific antibody generated in this study was used to assess the accumulation of the ycf4 gene product.
Figure 6.
Unaltered gene expression of all essential plastid-encoded PSI genes in Δ_ycf4_ knockout mutants. Translational efficiency for the essential plastid genome-encoded PSI-related genes psaA/B, psaC, and ycf3 was analyzed by polysome-loading assays. Collected fractions from the Suc density gradients are numbered from top to bottom. Equal aliquots of extracted RNAs from all fractions were separated by denaturing agarose gel electrophoresis, blotted, and hybridized to gene-specific radiolabeled probes. The arrows at the top indicate the gradient in Suc density (from low to high). As a control, a sample was treated with the antibiotic puromycin to cause the dissociation of ribosomes from the mRNAs. Ribosome distribution in the gradients is revealed by ethidium bromide (EtBr) staining of the agarose gels prior to blotting. PSI transcript patterns and mRNA accumulation levels were also determined (middle panel) for the three essential plastid genome-encoded PSI subunits (psaA/B and psaC) and the ycf3 gene encoding an essential PSI assembly factor (Ruf et al., 1997). To confirm equal loading, an ethidium bromide-stained agarose gels is also shown. WT, Wild type.
Figure 7.
Detection of Ycf4-containing high-M_r complexes by BN gel electrophoresis. A, One-dimensional BN gel electrophoretic separation of thylakoidal protein complexes. Transplastomic Δ_ycf4 knockout mutants, wild-type plants (WT), and KD-psaA control plants were analyzed. Arrows denote Ycf4-containing fractions. The asterisk indicates a nonspecific immunological cross-reaction (as evidenced by the presence of the signal in the Δ_ycf4_ knockout mutants). D, Dimer; M, monomer; SC, supercomplex; T, trimer. B, Two-dimensional electrophoretic analysis of PSI complexes and Ycf4-containing complexes in wild-type and Δ_ycf4_ transplastomic knockout plants. The first dimension was BN gel electrophoresis, and the second dimension was SDS-PAGE (see “Materials and Methods”). Nonspecific cross-reactions are indicated by the asterisk, and the Ycf4 protein is indicated by an arrow. C, Two-dimensional electrophoretic analysis of residual PSI complexes accumulating in the Δ_ycf4_ knockout mutant. The gels were blotted, and the blots were probed with antibodies against the PSI reaction center protein PsaA and the small PSI subunit PsaG. The identical electrophoretic migration of the PSI complexes from wild-type and mutant plants and the stable association of the small G-subunit with the complex also in the Δ_ycf4_ knockout suggest that the composition of the PSI complexes in the mutant is unaltered compared with the wild type.
Figure 8.
Developmental regulation of PSI assembly factors. Transplastomic Ycf3-FLAG lines (Albus et al., 2010) were used to follow the fate of three assembly factors for PSI: the plastid genome-encoded proteins Ycf3 and Ycf4 and the nuclear genome-encoded interaction partner of Ycf3, Y3IP1 (Schöttler et al., 2011). A developmental series of leaves from a tobacco plant prior to flowering is shown. Leaves are numbered from bottom to top. The two youngest leaves (nos. 10 and 11) had to be combined due to their small size. The assembly factors were detected with specific antibodies (for details, see text). For comparison, PSI accumulation was followed with an antibody against the essential reaction center subunit PsaA.
Similar articles
- Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in photosystem I assembly of tobacco and Arabidopsis.
Albus CA, Ruf S, Schöttler MA, Lein W, Kehr J, Bock R. Albus CA, et al. Plant Cell. 2010 Aug;22(8):2838-55. doi: 10.1105/tpc.110.073908. Epub 2010 Aug 31. Plant Cell. 2010. PMID: 20807881 Free PMC article. - Effects of site-directed mutations in the chloroplast-encoded Ycf4 gene on PSI complex assembly in the green alga Chlamydomonas reinhardtii.
Onishi T, Takahashi Y. Onishi T, et al. Plant Cell Physiol. 2009 Oct;50(10):1750-60. doi: 10.1093/pcp/pcp117. Epub 2009 Aug 10. Plant Cell Physiol. 2009. PMID: 19667102 - The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex.
Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix JD. Boudreau E, et al. EMBO J. 1997 Oct 15;16(20):6095-104. doi: 10.1093/emboj/16.20.6095. EMBO J. 1997. PMID: 9321389 Free PMC article. - Photosystem I: its biogenesis and function in higher plants.
Schöttler MA, Albus CA, Bock R. Schöttler MA, et al. J Plant Physiol. 2011 Aug 15;168(12):1452-61. doi: 10.1016/j.jplph.2010.12.009. Epub 2011 Jan 21. J Plant Physiol. 2011. PMID: 21255865 Review. - Tools and techniques for chloroplast transformation of Chlamydomonas.
Purton S. Purton S. Adv Exp Med Biol. 2007;616:34-45. doi: 10.1007/978-0-387-75532-8_4. Adv Exp Med Biol. 2007. PMID: 18161489 Review.
Cited by
- Multiple RNA processing defects and impaired chloroplast function in plants deficient in the organellar protein-only RNase P enzyme.
Zhou W, Karcher D, Fischer A, Maximova E, Walther D, Bock R. Zhou W, et al. PLoS One. 2015 Mar 20;10(3):e0120533. doi: 10.1371/journal.pone.0120533. eCollection 2015. PLoS One. 2015. PMID: 25793367 Free PMC article. - Tetratricopeptide repeat protein protects photosystem I from oxidative disruption during assembly.
Heinnickel M, Kim RG, Wittkopp TM, Yang W, Walters KA, Herbert SK, Grossman AR. Heinnickel M, et al. Proc Natl Acad Sci U S A. 2016 Mar 8;113(10):2774-9. doi: 10.1073/pnas.1524040113. Epub 2016 Feb 22. Proc Natl Acad Sci U S A. 2016. PMID: 26903622 Free PMC article. - Comparative Analyses of Plastomes of Four Anubias (Araceae) Taxa, Tropical Aquatic Plants Endemic to Africa.
Li L, Liu C, Hou K, Liu W. Li L, et al. Genes (Basel). 2022 Nov 5;13(11):2043. doi: 10.3390/genes13112043. Genes (Basel). 2022. PMID: 36360280 Free PMC article. - Complete plastome sequences from Glycine syndetika and six additional perennial wild relatives of soybean.
Sherman-Broyles S, Bombarely A, Grimwood J, Schmutz J, Doyle J. Sherman-Broyles S, et al. G3 (Bethesda). 2014 Aug 25;4(10):2023-33. doi: 10.1534/g3.114.012690. G3 (Bethesda). 2014. PMID: 25155272 Free PMC article. - Temporal Proteomics of Inducible RNAi Lines of Clp Protease Subunits Identifies Putative Protease Substrates.
Moreno JC, Martínez-Jaime S, Schwartzmann J, Karcher D, Tillich M, Graf A, Bock R. Moreno JC, et al. Plant Physiol. 2018 Feb;176(2):1485-1508. doi: 10.1104/pp.17.01635. Epub 2017 Dec 11. Plant Physiol. 2018. PMID: 29229697 Free PMC article.
References
- Amunts A, Drory O, Nelson N. (2007) The structure of a plant photosystem I supercomplex at 3.4 A resolution. Nature 447: 58–63 - PubMed
- Ayliffe MA, Scott NS, Timmis JN. (1998) Analysis of plastid DNA-like sequences within the nuclear genomes of higher plants. Mol Biol Evol 15: 738–745 - PubMed
- Bock R. (2001) Transgenic plastids in basic research and plant biotechnology. J Mol Biol 312: 425–438 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources