Structural basis of the intracellular sorting of the SNARE VAMP7 by the AP3 adaptor complex - PubMed (original) (raw)
Structural basis of the intracellular sorting of the SNARE VAMP7 by the AP3 adaptor complex
Helen M Kent et al. Dev Cell. 2012.
Abstract
VAMP7 is involved in the fusion of late endocytic compartments with other membranes. One possible mechanism of VAMP7 delivery to these late compartments is via the AP3 trafficking adaptor. We show that the linker of the δ-adaptin subunit of AP3 binds the VAMP7 longin domain and determines the structure of their complex. Mutation of residues on both partners abolishes the interaction in vitro and in vivo. The binding of VAMP7 to δ-adaptin requires the VAMP7 SNARE motif to be engaged in SNARE complex formation and hence AP3 must transport VAMP7 when VAMP7 is part of a cis-SNARE complex. The absence of δ-adaptin causes destabilization of the AP3 complex in mouse mocha fibroblasts and mislocalization of VAMP7. The mislocalization can be rescued by transfection with wild-type δ-adaptin but not by δ-adaptin containing mutations that abolish VAMP7 binding, despite in all cases intact AP3 being present and LAMP1 trafficking being rescued.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
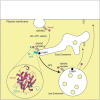
Graphical abstract
Figure 1
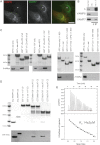
The δ-Adaptin Linker Binds to the Longin Domain Only of VAMP7 (A) Mocha fibroblasts were mixed with cells expressing δwt adaptin and then fixed and stained with antibodies to δ-adaptin and VAMP7. Asterisk indicates nontransduced cell. Scale bar, 20 μm. (B) Mocha fibroblasts were retrovirally transduced with δwt adaptin and native immunoprecipitations performed with antibodies to δ-adaptin. The immunoprecipitations were immunoblotted for the δ subunit of the AP-3 complex and the post-Golgi R-SNAREs VAMP8 (negative control) and VAMP7. As a positive control for the immunoblotting 3% of the mocha + δwt detergent extract was loaded (control). (C) In GST pull-down experiments, GSTVAMP7 longin domain (LD) but not cytoplasmic domain (CD) or GST α-ear bind to recombinant AP3 containing either full-length δ-adaptin (residues 1–1,203) or δ-adaptin trunk+hinge (residues 1–797) but not to AP3 containing δ-adaptin trunk (residues 1–620) or the δ-adaptin appendage (residues 720–1203). Top panels Coomassie blue staining and bottom panels western blots for the Myc tag at the C terminus of μ3 adaptin. (D) GST δ-adaptin (650–797) and GST δ-adaptin (680–728) but not GSTα-ear bind to His6MycVAMP7LD but not His6MycVAMPCD as detected by western blotting with an anti-Myc antibody. Top panels Coomassie blue staining and bottom panels western blots for the Myc tag on VAMP7 constructs. (E) VAMP7LD binds to GSTδ-adaptin (680–728) with a KD of 14 ± 2 μM by isothermal titration calorimetry. See also Figure S1.
Figure 2
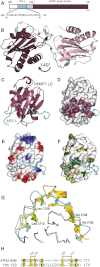
Structure of the δ-Adaptin Linker:VAMP7 Longin Domain Complex (A) Schematic representation of the δ-adaptin linker:VAMP7 longin domain fusion construct. As in all subsequent figures, the VAMP7 longin domain is shown in purple and the region of the δ-adaptin linker visible in the structure in cyan. The sequence of the δ-adaptin fragment is shown underneath. The numbers in italics correspond to the actual residue number in δ-adaptin, whereas those in regular font correspond to the numbers of the residues as they occur in the construct. (B) Structure of the linker exchanged dimer in the crystal's asymmetric unit. (C) Path followed by a δ-adaptin linker on a longin domain. (D) As (C) but with the longin domain shown in a gray surface representation with residues contacting the δ-adaptin linker colored purple. (E and F) As (C) but with longin domain surface colored by electrostatic potential (E) and hydrophobicity (ramped from polar (white) to hydrophobic (green)) (F) showing that the δ-adaptin linker sits in a hydrophobic groove. (G) Superposition of VAMP7:δ-adaptin linker (purple and cyan) and VAMP7:Hrb (gray and gold). Residues in Hrb (gold) spatially equivalent to those in δ-adaptin (green) mutation of which abolish the δ-adaptin/VAMP7 longin domain interaction are highlighted. (H) Structure based alignment of the δ-adaptin and Hrb fragments that bind the VAMP7 longin domain. Residues in bold mediate the interactions. Residues mutated in this studied are numbered. See also Figure S2.
Figure 3
Molecular Mechanism of the Interaction between the δ-Adaptin Linker and VAMP7 Longin Domain (A) View into the δ-adaptin (cyan) binding slot on the VAMP7 longin domain (purple). Residues whose mutation abolish the δ-adaptin/VAMP longin domain interaction are colored green (δ-adaptin) and magenta (VAMP7 longin domain). (B) In the context of GSTδ-adaptin hinge (residues 680–728), mutations L709S/L713S and I702S/V704S reduced binding to VAMP longin domain in pull-down experiments by 95% and 92% respectively as compared with binding to GSTδ-adaptin wt (680–728), and GSTδ-adaptin (680–728)V725S/M727S. Top panel Coomassie blue staining and bottom panel western blot with anti-VAMP7 antibody. Blot quantified with ImageJ software with amount of binding to the negative control (GSTα-ear) being subtracted from all lanes. (C) In the context of recombinant AP3 containing full-length δ-adaptin, wt δ-adaptin binds to GST VAMP7 longin domain in GST pull-down experiments whereas mutations L709S/L713S and I702S/V704S in δ-adaptin abolished the interaction. Top panel Coomassie blue staining and bottom panel western blot with anti-Myc antibody. (D) GST VAMP7 longin domain (LD) but not mutant GST VAMP7 (L43S/Y45S) or GSTαear bind to AP3 containing full-length δ-adaptin and Myc-tagged μ3 subunit. Top panel Coomassie blue staining and bottom panel western blot with anti-Myc antibody. Lane 2 is loaded with AP3 only. (E) GST VAMP7 longin domain (LD) but not GST VAMP7 cytosolic domain (CD) or GSTαear bind to AP3 containing Myc-tagged μ3 subunit. Formation of an endosomal SNARE complex on VAMP7CD confers on VAMP7CD the ability to bind AP3. Top panel Coomassie blue staining and bottom panel western blot with anti-Myc antibody. Lane 2 is loaded with AP3 only. (F) Schematic representation of the binding of a _cis_-SNARE complex containing VAMP7 by AP3 through the VAMP7's longin domain colored as in (A). See also Figure S3.
Figure 4
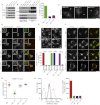
The Interaction between VAMP7 and AP3 Is Required for VAMP7's Steady-State Localization but Not Lysosomal Protein Targeting (A) Mocha fibroblasts were retrovirally transduced with δwt, δmut1, and δmut2 adaptin and levels of the AP3 subunits determined by immunoblotting. γ-Adaptin a subunit of the AP-1 adaptor complex and was used as a loading control. (B) Mocha fibroblasts were retrovirally transduced either with δwt, δmut1, and δmut2 adaptin and native immunoprecipitations performed with antibodies to δ-adaptin. The immunoprecipitations were immunoblotted for components of the AP-3 complex (μ3A and δ) and the post-Golgi R-SNAREs VAMP3 (negative control) and VAMP7. Immunoprecipitations were performed 3 times and the level of VAMP7 bound to AP-3 quantified (see graph). Error bars show the SEM. (C) Mocha fibroblasts were mixed with cells either expressing δwt, δmut1, and δmut2 adaptin and then fixed and stained with antibodies to δ-adaptin. Asterisk indicates nontransduced cell. Scale bar, 20 μm. (D) Mocha fibroblasts expressing δwt, δmut1, and δmut2 were transfected with GTP locked GFP-Rab5A. The cells were then fixed and stained with antibodies against δ adaptin. Scale bar 20 μm. (E) Mocha fibroblasts expressing δwt, δmut1, and δmut2 adaptin were fixed and stained with antibodies VAMP7. The micrographs show representative fields of cells. To quantify the localization of VAMP7, over 10, random micrographs were taken for each cell population and the localization of VAMP7 scored manually. The experiment was repeated 3 times and the error bars show the SEM n = total number of cells scored in the three independent experiments. Scale bar, 10 μm. (F) Mocha fibroblasts expressing δwt, δmut1, and δmut2 were fixed and stained with antibodies to VAMP7 and vti1a. Scale bar 20 μm. (G) The level of colocalization between VAMP7 and vti1a was calculated using Volocity software. The bar graphs show mean Pearson coefficients and the error bars SEM 10 cells were imaged for each condition. (H) Mocha fibroblasts reterovirally transduced either with δwt, δmut1, and δmut2 adaptin and the cell surface levels of LAMP1 determined using flow cytometry. The raw data from a representative experiment is shown in the histogram (black line unstained-control, red-mh, green-mh + δwt, blue-mh + δmut1 and magenta-mh + δmut2). The bar graph show the data generated from three independent experiments. Error bars show experimental range. See also Figures S4 and S5.
Similar articles
- Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb.
Pryor PR, Jackson L, Gray SR, Edeling MA, Thompson A, Sanderson CM, Evans PR, Owen DJ, Luzio JP. Pryor PR, et al. Cell. 2008 Sep 5;134(5):817-27. doi: 10.1016/j.cell.2008.07.023. Cell. 2008. PMID: 18775314 Free PMC article. - Sorting of the v-SNARE VAMP7 in Dictyostelium discoideum: a role for more than one Adaptor Protein (AP) complex.
Bennett N, Letourneur F, Ragno M, Louwagie M. Bennett N, et al. Exp Cell Res. 2008 Sep 10;314(15):2822-33. doi: 10.1016/j.yexcr.2008.06.019. Epub 2008 Jul 2. Exp Cell Res. 2008. PMID: 18634783 - Identification of the yeast R-SNARE Nyv1p as a novel longin domain-containing protein.
Wen W, Chen L, Wu H, Sun X, Zhang M, Banfield DK. Wen W, et al. Mol Biol Cell. 2006 Oct;17(10):4282-99. doi: 10.1091/mbc.e06-02-0128. Epub 2006 Jul 19. Mol Biol Cell. 2006. PMID: 16855025 Free PMC article. - Multiple Roles of VARP in Endosomal Trafficking: Rabs, Retromer Components and R-SNARE VAMP7 Meet on VARP.
Fukuda M. Fukuda M. Traffic. 2016 Jul;17(7):709-19. doi: 10.1111/tra.12406. Epub 2016 May 13. Traffic. 2016. PMID: 27103185 Review. - The delivery of endocytosed cargo to lysosomes.
Luzio JP, Parkinson MD, Gray SR, Bright NA. Luzio JP, et al. Biochem Soc Trans. 2009 Oct;37(Pt 5):1019-21. doi: 10.1042/BST0371019. Biochem Soc Trans. 2009. PMID: 19754443 Review.
Cited by
- Increased activity of the vesicular soluble N-ethylmaleimide-sensitive factor attachment protein receptor TI-VAMP/VAMP7 by tyrosine phosphorylation in the Longin domain.
Burgo A, Casano AM, Kuster A, Arold ST, Wang G, Nola S, Verraes A, Dingli F, Loew D, Galli T. Burgo A, et al. J Biol Chem. 2013 Apr 26;288(17):11960-72. doi: 10.1074/jbc.M112.415075. Epub 2013 Mar 7. J Biol Chem. 2013. PMID: 23471971 Free PMC article. - VAMP7 modulates ciliary biogenesis in kidney cells.
Szalinski CM, Labilloy A, Bruns JR, Weisz OA. Szalinski CM, et al. PLoS One. 2014 Jan 22;9(1):e86425. doi: 10.1371/journal.pone.0086425. eCollection 2014. PLoS One. 2014. PMID: 24466086 Free PMC article. - Amyloid-like aggregating proteins cause lysosomal defects in neurons via gain-of-function toxicity.
Riera-Tur I, Schäfer T, Hornburg D, Mishra A, da Silva Padilha M, Fernández-Mosquera L, Feigenbutz D, Auer P, Mann M, Baumeister W, Klein R, Meissner F, Raimundo N, Fernández-Busnadiego R, Dudanova I. Riera-Tur I, et al. Life Sci Alliance. 2021 Dec 21;5(3):e202101185. doi: 10.26508/lsa.202101185. Print 2022 Mar. Life Sci Alliance. 2021. PMID: 34933920 Free PMC article. - The Sybtraps: control of synaptobrevin traffic by synaptophysin, α-synuclein and AP-180.
Gordon SL, Cousin MA. Gordon SL, et al. Traffic. 2014 Mar;15(3):245-54. doi: 10.1111/tra.12140. Epub 2013 Dec 16. Traffic. 2014. PMID: 24279465 Free PMC article. Review. - Presynaptic membrane retrieval and endosome biology: defining molecularly heterogeneous synaptic vesicles.
Morgan JR, Comstra HS, Cohen M, Faundez V. Morgan JR, et al. Cold Spring Harb Perspect Biol. 2013 Oct 1;5(10):a016915. doi: 10.1101/cshperspect.a016915. Cold Spring Harb Perspect Biol. 2013. PMID: 24086045 Free PMC article. Review.
References
- Advani R.J., Bae H.R., Bock J.B., Chao D.S., Doung Y.C., Prekeris R., Yoo J.S., Scheller R.H. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J. Biol. Chem. 1998;273:10317–10324. - PubMed
- Chaineau M., Danglot L., Galli T. Multiple roles of the vesicular-SNARE TI-VAMP in post-Golgi and endosomal trafficking. FEBS Lett. 2009;583:3817–3826. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 079895/WT_/Wellcome Trust/United Kingdom
- G120/952/MRC_/Medical Research Council/United Kingdom
- MC_U105178845/MRC_/Medical Research Council/United Kingdom
- U105178845/MRC_/Medical Research Council/United Kingdom
- 090909/WT_/Wellcome Trust/United Kingdom
- G0900113/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous