A PPARγ-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis - PubMed (original) (raw)
. 2012 May 17;485(7398):391-4.
doi: 10.1038/nature10998.
Jae Myoung Suh, Annette R Atkins, Maryam Ahmadian, Pingping Li, Jamie Whyte, Mingxiao He, Henry Juguilon, Yun-Qiang Yin, Colin T Phillips, Ruth T Yu, Jerrold M Olefsky, Robert R Henry, Michael Downes, Ronald M Evans
Affiliations
- PMID: 22522926
- PMCID: PMC3358516
- DOI: 10.1038/nature10998
A PPARγ-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis
Johan W Jonker et al. Nature. 2012.
Abstract
Although feast and famine cycles illustrate that remodelling of adipose tissue in response to fluctuations in nutrient availability is essential for maintaining metabolic homeostasis, the underlying mechanisms remain poorly understood. Here we identify fibroblast growth factor 1 (FGF1) as a critical transducer in this process in mice, and link its regulation to the nuclear receptor PPARγ (peroxisome proliferator activated receptor γ), which is the adipocyte master regulator and the target of the thiazolidinedione class of insulin sensitizing drugs. FGF1 is the prototype of the 22-member FGF family of proteins and has been implicated in a range of physiological processes, including development, wound healing and cardiovascular changes. Surprisingly, FGF1 knockout mice display no significant phenotype under standard laboratory conditions. We show that FGF1 is highly induced in adipose tissue in response to a high-fat diet and that mice lacking FGF1 develop an aggressive diabetic phenotype coupled to aberrant adipose expansion when challenged with a high-fat diet. Further analysis of adipose depots in FGF1-deficient mice revealed multiple histopathologies in the vasculature network, an accentuated inflammatory response, aberrant adipocyte size distribution and ectopic expression of pancreatic lipases. On withdrawal of the high-fat diet, this inflamed adipose tissue fails to properly resolve, resulting in extensive fat necrosis. In terms of mechanisms, we show that adipose induction of FGF1 in the fed state is regulated by PPARγ acting through an evolutionarily conserved promoter proximal PPAR response element within the FGF1 gene. The discovery of a phenotype for the FGF1 knockout mouse establishes the PPARγ–FGF1 axis as critical for maintaining metabolic homeostasis and insulin sensitization.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
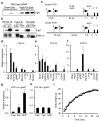
Figure 1. FGF1A is induced in adipose tissue by high-fat diet (HFD)
a, Western blot of FGF1 in gonadal white adipose (gWAT) of chow or HFD-fed wild-type mice (n=3). b, Western blot of FGF1 in whole fat, adipocyte (adip.) and stromal vascular fractions (SVF) of inguinal (ing.) and gonadal (gon.) white adipose of chow-fed wild-type mice (ERK1/2 loading control). c, Diagram depicting three distinct promoters driving the untranslated exons 1A, 1B, and 1D (open bars) of human and mouse FGF1 genes. Alternative splicing of untranslated exons results in identical FGF1 polypeptides. d–f, mRNA tissue distribution in mice for FGF1A (d), FGF1B (e), and FGF1D (f). g, h, mRNA levels of FGF1A (g) and FGF1B (h) in gWAT of chow fed, 2-weeks HFD-fed, or overnight fasted wild-type mice (n = 5). i, Body weight of HFD-fed wild-type and FGF1−/− mice over 24 weeks. Data expressed as mean ± S.D. * *p<0.01.
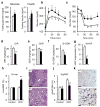
Figure 2. Loss of FGF1 results in diet-induced insulin resistance
Metabolic studies on 24 week old male wild-type (open bars) and FGF1−/− (filled bars) mice fed a HFD for 16 weeks. a, Fasting serum glucose and insulin levels. b, c, Glucose (b) and insulin (c) tolerance tests. d–g, Glucose infusion rate (GIR) (d), glucose disposal rate (GDR) (e), insulin-stimulated GDR (IS-GDR) (f), and percent suppression of hepatic glucose production (HGP) (g) during hyperinsulinemic-euglycemic clamp studies. h, Liver (percent body weight) from chow and HFD-fed mice (n=6–7). i, H&E staining of liver from HFD-fed wild-type (WT) and FGF1−/− (KO) mice. j, Gonadal white adipose (gWAT) (percent body weight) from chow and HFD-fed mice (n=6–7). k, H&E staining of gWAT from HFD-fed wild-type (WT) and FGF1−/− (KO) mice. Scale bar = 100 μm. Data expressed as mean ± S.D. *p<0.05, **p<0.01.
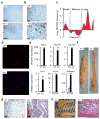
Figure 3. Loss of FGF1 results in defects in adipose remodelling during HFD
a, Immunohistochemistry for the macrophage marker, F4/80 in gWAT from HFD-fed wild-type (WT) and FGF1−/− (KO) mice. b, Trichrome staining for collagen deposition in gWAT from HFD-fed wild-type (WT) and FGF1−/− (KO) mice. c, Quantitation of adipose cell cross-sectional area from HFD-fed mice, expressed as the ratio of the number of cells from FGF1−/− (KO) to wild-type (WT) mice defined as small (1000–4000 μm2), medium (4000–10000 μm2) and large (>10000 μm2) cells. d, Fluorescence microscopy of gWAT from HFD-fed wild-type (WT) and FGF1−/− (KO) mice perfused with microbeads (red) and counterstained with DAPI (blue). e, QPCR verification of induction of genes associated with fat necrosis in HFD-fed wild-type (open bars) and FGF1−/− (closed bars) mice. f, gWAT depots from HFD to chow converted (HCC) wild-type (WT) and FGF1−/− (KO) mice. g, H&E staining of gWAT from HCC wild-type (WT) and FGF1−/− (KO) mice. h, Image and H&E staining of dissociated necrotic WAT taken from peritoneal cavity of HCC FGF1−/− (KO) mice. Scale bar = 100 μm. Data expressed as mean ± S.D. **p<0.01.
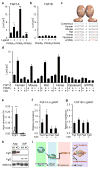
Figure 4. FGF1 is a direct transcriptional target of PPARγ
a, b, Luciferase reporter assays of FGF1A (a) and FGF1B (b) promoters co-transfected with PPARs +/− ligand. c, Sequence alignment of the putative PPRE within the FGF1A promoter from different species. Red indicates nucleotide variations between the PPREs relative to human. d, Species-specific response of the FGF1A promoters to PPARγ +/− ligand using luciferase reporter assays. e, Chromatin immuno-precipitation of PPARγ on the FGF1A promoter in differentiated 3T3-L1 cells (open bars IgG, closed bars PPARγ antibody). f, g, Levels of FGF1A (f) FGF1B (g) mRNA in gonadal white adipose (gWAT) of fed or overnight fasted wild-type mice (n=5) with or w/o TZD (5mg/kg for 3 days i.p). h, Western blot of FGF1, FGF2 in adipocyte (adip.) and stromal vascular (SVF) fractions of gonadal white adipose (gWAT) from chow fed wild-type (WT) and aP2-Cre; PPARγfl/fl (adipocyte-specific PPARγ knockout) mice (KO) (ERK1/2 loading control). i, Model depicting the role of the PPARγ-FGF1 axis in adipose remodelling and insulin sensitivity. Data expressed as mean ± S.D. *p<0.05,**p<0.01.
Similar articles
- The PPARγ-FGF1 axis: an unexpected mediator of adipose tissue homeostasis.
Sun K, Scherer PE. Sun K, et al. Cell Res. 2012 Oct;22(10):1416-8. doi: 10.1038/cr.2012.94. Epub 2012 Jun 19. Cell Res. 2012. PMID: 22710798 Free PMC article. - Peroxisome Proliferator-Activated Receptor γ and Its Role in Adipocyte Homeostasis and Thiazolidinedione-Mediated Insulin Sensitization.
Wang QA, Zhang F, Jiang L, Ye R, An Y, Shao M, Tao C, Gupta RK, Scherer PE. Wang QA, et al. Mol Cell Biol. 2018 Apr 30;38(10):e00677-17. doi: 10.1128/MCB.00677-17. Print 2018 May 15. Mol Cell Biol. 2018. PMID: 29483301 Free PMC article. - Artemisia extracts activate PPARγ, promote adipogenesis, and enhance insulin sensitivity in adipose tissue of obese mice.
Richard AJ, Burris TP, Sanchez-Infantes D, Wang Y, Ribnicky DM, Stephens JM. Richard AJ, et al. Nutrition. 2014 Jul-Aug;30(7-8 Suppl):S31-6. doi: 10.1016/j.nut.2014.02.013. Epub 2014 Mar 12. Nutrition. 2014. PMID: 24985103 Free PMC article. - FGF1.
Jamal SB, Hockman D. Jamal SB, et al. Differentiation. 2024 Sep-Oct;139:100802. doi: 10.1016/j.diff.2024.100802. Epub 2024 Jul 22. Differentiation. 2024. PMID: 39074995 Review. - Adipogenesis and lipotoxicity: role of peroxisome proliferator-activated receptor gamma (PPARgamma) and PPARgammacoactivator-1 (PGC1).
Medina-Gomez G, Gray S, Vidal-Puig A. Medina-Gomez G, et al. Public Health Nutr. 2007 Oct;10(10A):1132-7. doi: 10.1017/S1368980007000614. Public Health Nutr. 2007. PMID: 17903321 Review.
Cited by
- FGF1 and FGF19 reverse diabetes by suppression of the hypothalamic-pituitary-adrenal axis.
Perry RJ, Lee S, Ma L, Zhang D, Schlessinger J, Shulman GI. Perry RJ, et al. Nat Commun. 2015 Apr 28;6:6980. doi: 10.1038/ncomms7980. Nat Commun. 2015. PMID: 25916467 Free PMC article. - Monogenic Diabetes: What It Teaches Us on the Common Forms of Type 1 and Type 2 Diabetes.
Yang Y, Chan L. Yang Y, et al. Endocr Rev. 2016 Jun;37(3):190-222. doi: 10.1210/er.2015-1116. Epub 2016 Apr 1. Endocr Rev. 2016. PMID: 27035557 Free PMC article. Review. - PPARγ signaling and metabolism: the good, the bad and the future.
Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, Evans RM. Ahmadian M, et al. Nat Med. 2013 May;19(5):557-66. doi: 10.1038/nm.3159. Epub 2013 May 7. Nat Med. 2013. PMID: 23652116 Free PMC article. Review. - Targeting the brain as a cure for type 2 diabetes.
Seeley RJ, Sandoval DA. Seeley RJ, et al. Nat Med. 2016 Jul 7;22(7):709-11. doi: 10.1038/nm.4137. Nat Med. 2016. PMID: 27387884 No abstract available. - Adipose tissue plasticity from WAT to BAT and in between.
Lee YH, Mottillo EP, Granneman JG. Lee YH, et al. Biochim Biophys Acta. 2014 Mar;1842(3):358-69. doi: 10.1016/j.bbadis.2013.05.011. Epub 2013 May 17. Biochim Biophys Acta. 2014. PMID: 23688783 Free PMC article. Review.
References
- Forman BM, et al. 15-Deoxy-Δ12,14-Prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell. 1995;83:803–812. - PubMed
- Barak Y, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. - PubMed
- Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. - PubMed
References in methods
- Hevener AL, et al. Muscle-specific Pparg deletion causes insulin resistance. Nat Med. 2003;9:1491–1497. - PubMed
- Springer ML, Ip TK, Blau HM. Angiogenesis monitored by perfusion with a space-filling microbead suspension. Mol Ther. 2000;1:82–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R24 DK090962-02/DK/NIDDK NIH HHS/United States
- R01 HL105278/HL/NHLBI NIH HHS/United States
- R37 DK057978-34/DK/NIDDK NIH HHS/United States
- DK062434/DK/NIDDK NIH HHS/United States
- R01 HL105278-21/HL/NHLBI NIH HHS/United States
- U19 DK062434/DK/NIDDK NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- U19 DK062434-10/DK/NIDDK NIH HHS/United States
- R37 DK057978/DK/NIDDK NIH HHS/United States
- DK063491/DK/NIDDK NIH HHS/United States
- HL105278/HL/NHLBI NIH HHS/United States
- P50 HD012303/HD/NICHD NIH HHS/United States
- DK057978/DK/NIDDK NIH HHS/United States
- R24 DK090962/DK/NIDDK NIH HHS/United States
- R01 DK033651/DK/NIDDK NIH HHS/United States
- DK090962/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R37 DK033651/DK/NIDDK NIH HHS/United States
- R01 DK057978/DK/NIDDK NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases