Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment - PubMed (original) (raw)
. 2012 May 8;109(19):7362-7.
doi: 10.1073/pnas.1201595109. Epub 2012 Apr 23.
Biswarup Saha, Soma Ray, Debasree Dutta, Sumedha Gunewardena, Byunggil Yoo, Arindam Pal, Jay L Vivian, Melissa Larson, Margaret Petroff, Patrick G Gallagher, Vincent P Schulz, Kenneth L White, Thaddeus G Golos, Barry Behr, Soumen Paul
Affiliations
- PMID: 22529382
- PMCID: PMC3358889
- DOI: 10.1073/pnas.1201595109
Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment
Pratik Home et al. Proc Natl Acad Sci U S A. 2012.
Abstract
In the preimplantation mouse embryo, TEAD4 is critical to establishing the trophectoderm (TE)-specific transcriptional program and segregating TE from the inner cell mass (ICM). However, TEAD4 is expressed in the TE and the ICM. Thus, differential function of TEAD4 rather than expression itself regulates specification of the first two cell lineages. We used ChIP sequencing to define genomewide TEAD4 target genes and asked how transcription of TEAD4 target genes is specifically maintained in the TE. Our analyses revealed an evolutionarily conserved mechanism, in which lack of nuclear localization of TEAD4 impairs the TE-specific transcriptional program in inner blastomeres, thereby allowing their maturation toward the ICM lineage. Restoration of TEAD4 nuclear localization maintains the TE-specific transcriptional program in the inner blastomeres and prevents segregation of the TE and ICM lineages and blastocyst formation. We propose that altered subcellular localization of TEAD4 in blastomeres dictates first mammalian cell fate specification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
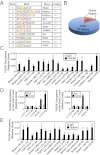
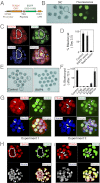
Direct regulation of core TSC genes by TEAD4. (A) HOMER analysis shows most abundant transcription factor-binding motifs at the ChIP-seq–identified TEAD4 binding regions along with log P values. (B) Pie chart shows numbers of core mTSC-specific genes with identified TEAD4 binding regions within ±50 kb of the transcription start site. (C) Quantitative ChIP analyses (mean ± SE; n = 3) shows TEAD4 chromatin occupancy at 18 regions that were identified by ChIP-seq analysis. The red bar shows a region in which TEAD4 occupancy was not detected. (D) ChIP analysis shows TEAD4 occupancy (mean ± SE; n = 3) at Gata3 and Cdx2 loci in mTSCs. (E) ChIP analyses (mean ± SE; n = 3) in mouse blastocysts show TEAD4 occupancy at the chromatin domains of mTSC-specific genes, including Gata3 and Cdx2.
Fig. 2.
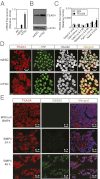
TEAD4 nuclear localization is impaired in ESCs. Quantitative RT-PCR (A) and Western blot (B) analyses of TEAD4 expression in undifferentiated mESCs and mTSCs. (C) Quantitative ChIP analyses for TEAD4 occupancy at its target loci in undifferentiated mESCs (mean ± SE; n = 3). (D) Confocal images of undifferentiated mESCs and mTSCs show cellular localization of TEAD4 with respect to nuclei. (E) H9 hESCs were treated with BMP4 for different time intervals, and confocal images were taken to show that TEAD4 protein (red) localization in the nuclei (blue) precedes GATA3 (green) expression.
Fig. 3.
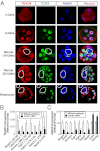
Impaired nuclear localization of TEAD4 represses target genes in the inner blastomeres of a preimplantation mouse embryo. (A) Preimplantation mouse embryos at different developmental stages were analyzed by confocal microscopy for TEAD4 (red) and CDX2 (green) expression with respect to nuclei (blue). Three-dimensional projections of confocal images at morula-stage embryos demonstrate lack of TEAD4 nuclear localization and CDX2 expression in the inner cells (white border). The white arrows show a dividing cell. Bottom: Micrographs show confocal images of blastocysts with near-complete exclusion of TEAD4 and loss of CDX2 expression in the nuclei of the ICM (white borders). Quantitative ChIP (B) and RT-PCR (C) analyses show decreased TEAD4 chromatin occupancy at its target loci, and repression of TEAD4 target genes in the inner cells of late morula/early blastocyst-stage mouse embryos compared with whole embryos (mean ± SE; n = 3). Note high levels of Oct4 and Nanog mRNA expression in the inner cells.
Fig. 4.
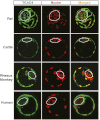
Impaired TEAD4 nuclear localization in the ICM lineages is a conserved phenomenon during mammalian preimplantation development. Immunofluorescence analyses show localization of TEAD4 protein (green) with respect to nuclei (red) in rat, cattle, monkey, and human blastocysts as observed by confocal microscopy.
Fig. 5.
Induced TEAD4 nuclear localization in the inner blastomeres impairs preimplantation mouse development. (A) Schematic diagram of the _Tead4_-_T2A_-EGFP lentiviral construct. (B) One-cell mouse embryos were infected with the _Tead4_-_T2A_-EGFP lentiviral particles and were cultured for 96 h. Differential interference contrast (Left) and fluorescence (Right) micrographs show that development of embryos with ectopic TEAD4 expression (detected by EGFP expression) was arrested at the morula stage. A control embryo without lentiviral infection (black arrow, differential interference contrast micrograph) developed to the late blastocyst stage when cultured for the same period. (C) Three-dimension projection of confocal images of a TEAD4-overexpressing embryo. Presence of TEAD4 and CDX2 in all the nuclei is indicated with white borders. (D) Graph shows percentage of embryos maturing to the blastocyst stage after 96 h of culture. (E) One-cell mouse embryos were treated with or without BMP4, and preimplantation embryonic development was monitored. Micrographs show that, after 72 h, most embryos without BMP4 treatment are maturing to the blastocyst stage; BMP4 treatment arrested embryos at the morula stage. (F) One-cell mouse embryos were cultured for 72 h and treated with BMP4 at different developmental stages. The graph shows the percentage of embryos maturing to the blastocyst stage. (G) Three-dimensional projections of confocal images of morula-stage embryos treated with BMP4 from the four-cell stage. Two different embryos from two independent experiments are shown. The images show presence of TEAD4 and CDX2 in inner cell nuclei (white arrows). (H) Confocal images of a vehicle-injected blastocyst shows a lack of TEAD4 nuclear localization and CDX2 expression in the ICM. (I) Three-dimensional projection of confocal images of a BMP4-injected embryo shows nuclear TEAD4 and CDX2 expression in the ICM cells (white arrows).
Fig. 6.
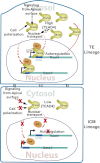
TEAD4 mediated specification of TE and ICM lineages in a preimplantation mammalian embryo. The model illustrates that nuclear localization of TEAD4 in the outer TE lineage induces TE-specific genes like Gata3. TEAD4 nuclear localization positively autoregulates its own transcription and increases TEAD4 protein levels in the TE lineage. The high TEAD4 concentration facilitates its nuclear localization. The dotted line denotes other putative mechanisms, not yet supported by experimental evidence, which could regulate TEAD4 nuclear localization in polar outer cells. The model also predicts that low TEAD4, as well as the absence of other putative mechanisms, impairs TEAD4 nuclear localization in the ICM lineage, thereby limiting TEAD4 transcription and abrogating expression of other TE-specific genes, such as Gata3.
Comment in
- Tead4 is constitutively nuclear, while nuclear vs. cytoplasmic Yap distribution is regulated in preimplantation mouse embryos.
Hirate Y, Cockburn K, Rossant J, Sasaki H. Hirate Y, et al. Proc Natl Acad Sci U S A. 2012 Dec 11;109(50):E3389-90; author reply E3391-2. doi: 10.1073/pnas.1211810109. Epub 2012 Nov 20. Proc Natl Acad Sci U S A. 2012. PMID: 23169672 Free PMC article. No abstract available.
Similar articles
- TEAD4 regulates trophectoderm differentiation upstream of CDX2 in a GATA3-independent manner in the human preimplantation embryo.
Stamatiadis P, Cosemans G, Boel A, Menten B, De Sutter P, Stoop D, Chuva de Sousa Lopes SM, Lluis F, Coucke P, Heindryckx B. Stamatiadis P, et al. Hum Reprod. 2022 Jul 30;37(8):1760-1773. doi: 10.1093/humrep/deac138. Hum Reprod. 2022. PMID: 35700449 - Changes in the expression patterns of the genes involved in the segregation and function of inner cell mass and trophectoderm lineages during porcine preimplantation development.
Fujii T, Sakurai N, Osaki T, Iwagami G, Hirayama H, Minamihashi A, Hashizume T, Sawai K. Fujii T, et al. J Reprod Dev. 2013;59(2):151-8. doi: 10.1262/jrd.2012-122. Epub 2012 Dec 20. J Reprod Dev. 2013. PMID: 23257836 Free PMC article. - Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos.
Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Nishioka N, et al. Mech Dev. 2008 Mar-Apr;125(3-4):270-83. doi: 10.1016/j.mod.2007.11.002. Epub 2007 Nov 17. Mech Dev. 2008. PMID: 18083014 - Mechanisms of trophectoderm fate specification in preimplantation mouse development.
Sasaki H. Sasaki H. Dev Growth Differ. 2010 Apr;52(3):263-73. doi: 10.1111/j.1440-169X.2009.01158.x. Epub 2010 Jan 20. Dev Growth Differ. 2010. PMID: 20100249 Review. - Establishment of trophectoderm and inner cell mass lineages in the mouse embryo.
Marikawa Y, Alarcón VB. Marikawa Y, et al. Mol Reprod Dev. 2009 Nov;76(11):1019-32. doi: 10.1002/mrd.21057. Mol Reprod Dev. 2009. PMID: 19479991 Free PMC article. Review.
Cited by
- Role of Hippo signaling pathway in early placental development.
Soncin F, Parast MM. Soncin F, et al. Proc Natl Acad Sci U S A. 2020 Aug 25;117(34):20354-20356. doi: 10.1073/pnas.2013559117. Epub 2020 Aug 11. Proc Natl Acad Sci U S A. 2020. PMID: 32788372 Free PMC article. No abstract available. - Atypical protein kinase C iota (PKCλ/ι) ensures mammalian development by establishing the maternal-fetal exchange interface.
Bhattacharya B, Home P, Ganguly A, Ray S, Ghosh A, Islam MR, French V, Marsh C, Gunewardena S, Okae H, Arima T, Paul S. Bhattacharya B, et al. Proc Natl Acad Sci U S A. 2020 Jun 23;117(25):14280-14291. doi: 10.1073/pnas.1920201117. Epub 2020 Jun 8. Proc Natl Acad Sci U S A. 2020. PMID: 32513715 Free PMC article. - Review: Trophoblast differentiation from human embryonic stem cells.
Golos TG, Giakoumopoulos M, Gerami-Naini B. Golos TG, et al. Placenta. 2013 Mar;34 Suppl:S56-61. doi: 10.1016/j.placenta.2012.11.019. Epub 2012 Dec 20. Placenta. 2013. PMID: 23261342 Free PMC article. Review. - Regulation of energy metabolism during early mammalian development: TEAD4 controls mitochondrial transcription.
Kumar RP, Ray S, Home P, Saha B, Bhattacharya B, Wilkins HM, Chavan H, Ganguly A, Milano-Foster J, Paul A, Krishnamurthy P, Swerdlow RH, Paul S. Kumar RP, et al. Development. 2018 Oct 1;145(19):dev162644. doi: 10.1242/dev.162644. Development. 2018. PMID: 30201685 Free PMC article.
References
- Zernicka-Goetz M. Cleavage pattern and emerging asymmetry of the mouse embryo. Nat Rev Mol Cell Biol. 2005;6:919–928. - PubMed
- Yagi R, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. - PubMed
- Ralston A, et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. - PubMed
- Nishioka N, et al. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125:270–283. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL094892/HL/NHLBI NIH HHS/United States
- R01 HD045611/HD/NICHD NIH HHS/United States
- R01 RR021876/RR/NCRR NIH HHS/United States
- R21 HL106311/HL/NHLBI NIH HHS/United States
- R01 HD062546/HD/NICHD NIH HHS/United States
- R21 HL094892/HL/NHLBI NIH HHS/United States
- HL106311/HL/NHLBI NIH HHS/United States
- HD53925/HD/NICHD NIH HHS/United States
- RR000167/RR/NCRR NIH HHS/United States
- RR21876/RR/NCRR NIH HHS/United States
- P51 OD011106/OD/NIH HHS/United States
- HD062546/HD/NICHD NIH HHS/United States
- P51 RR000167/RR/NCRR NIH HHS/United States
- R21 HD053925/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases