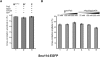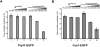Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS - PubMed (original) (raw)
Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS
Thomas Ohrt et al. RNA. 2012 Jun.
Abstract
The compositional and conformational changes during catalytic activation of the spliceosome promoted by the DEAH box ATPase Prp2 are only poorly understood. Here, we show by dual-color fluorescence cross-correlation spectroscopy (dcFCCS) that the binding affinity of several proteins is significantly changed during the Prp2-mediated transition of precatalytic B(act) spliceosomes to catalytically activated B* spliceosomes from Saccharomyces cerevisiae. During this step, several proteins, including the zinc-finger protein Cwc24, are quantitatively displaced from the B* complex. Consistent with this, we show that Cwc24 is required for step 1 but not for catalysis per se. The U2-associated SF3a and SF3b proteins Prp11 and Cus1 remain bound to the B* spliceosome under near-physiological conditions, but their binding is reduced at high salt. Conversely, high-affinity binding sites are created for Yju2 and Cwc25 during catalytic activation, consistent with their requirement for step 1 catalysis. Our results suggest high cooperativity of multiple Prp2-mediated structural rearrangements at the spliceosome's catalytic core. Moreover, dcFCCS represents a powerful tool ideally suited to study quantitatively spliceosomal protein dynamics in equilibrium.
Figures
FIGURE 1.
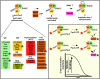
Experimental design and example of data. The Prp2-deficient spliceosomal complex BactΔPrp2 (stalled through its lack of Prp2) is catalytically activated by addition of Prp2 together with Spp2 and ATP. This leads to complex B*, which, on binding the step 1 factor Cwc25, catalyzes step 1 of splicing, generating complex C; for details, see Introduction. U2/5/6 snRNPs are shown as circles. The composition of the activated spliceosome is shown underneath; green stars indicate the spliceosomal proteins that were tagged with EGFP. (Inset) Release or recruitment of an EGFP-labeled protein (green star) from/to the spliceosome (carrying Atto647N, red star). The cross-correlation amplitude at short correlation time τ is proportional to the amount of doubly labeled spliceosomes.
FIGURE 2.
Binding behavior of Snu114-EGFP fusion protein to the spliceosome analyzed by dcFCCS. (A) Affinity-purified BactΔPrp2 complexes assembled on Atto647N-M3Act, carrying Snu114-EGFP (column 1), were complemented with Prp2, Spp2, and ATP (column 2) and Cwc25 (column 3). After incubation (45 min at 23°C; see Materials and Methods), dcFCCS measurements were then performed at complex concentrations of 1.0 nM. Cross-correlation amplitudes derived from two independent preparations are shown for each complex. Error bars indicate the standard deviation from two independent measurements. (B) Salt-resistance of Snu114-EGFP binding to the spliceosome during catalytic activation. Affinity-purified BactΔPrp2 complexes assembled on Atto647N-M3Act, carrying Snu114-EGFP (columns 1_–_3), were complemented with Prp2, Spp2, and ATP (columns 4_–_6). After the standard incubation, increasing concentrations of KCl were added to the samples, which were then subjected to dcFCCS measurements.
FIGURE 3.
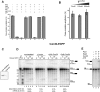
Displacement of Cwc24 from the spliceosome during Prp2-mediated catalytic activation, implying its noninvolvement in catalytic step 1 of splicing. (A) Affinity-purified BactΔPrp2 complexes assembled on Atto647N-M3Act, carrying Cwc24-EGFP (column 1), were complemented as indicated above each bar, or (B) were eluted from the affinity column and incubated with increasing concentrations of KCl for 30 min on ice before dcFCCS measurements, as shown. (C) Western blot analysis of yeast splicing extracts carrying Cwc24 tagged with the TAP tag before and after depletion of Cwc24 (ΔCwc24). (D) A uniformly 32P-labeled M3Act pre-mRNA was incubated in yeast whole-cell extract, which was either nondepleted (lanes 1_–_5) or Cwc24-depleted (lanes 6_–_20) under standard splicing conditions. Recombinant Cwc24 was then added to a final concentration of 1 or 3 μM. The splicing mixtures were incubated at 23°C and stopped at the time indicated. RNA was analyzed on an 8% urea–polyacrylamide gel and visualized by autoradiography. The positions of the pre-mRNAs, the splicing intermediates, and products are indicated on the right. The lariat-intron and the mRNA observed after 20-min incubation were quantified using Quantity One software (BioRad), and the results are shown below the gel. (E) Affinity-purified BactΔPrp2 complexes assembled on 32P-labeled M3Act, carrying Cwc24-EGFP (lane 1), were complemented with Prp2, Spp2, and ATP (lane 2), plus the addition of Cwc25 (lane 3). The positions of the pre-mRNAs, the splicing intermediates, and products are indicated on the left.
FIGURE 4.
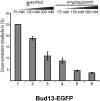
Displacement of Bud13-EGFP from the spliceosome during Prp2-mediated catalytic activation. Affinity-purified BactΔPrp2 complexes assembled on Atto647N-M3Act, carrying Bud13-EGFP (columns 1_–_3), were complemented with Prp2, Spp2, and ATP (columns 4_–_6). After incubation, KCl was added in increasing concentrations, and dcFCCS was performed as above.
FIGURE 5.
Binding of U2 SF3a/b proteins Prp11-EGFP and Cus1-EGFP to the spliceosome is made salt-sensitive by the catalytic activation. Affinity-purified BactΔPrp2 complexes were assembled on Atto647N-M3Act, carrying Prp11-EGFP (A) or Cus1-EGFP (B). The complexes were incubated with increasing salt concentration before (columns 1_–_3) or after (columns 4_–_6) complementation with Prp2, Spp2, and ATP.
FIGURE 6.
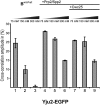
Strengthening of binding of Yju2-EGFP to the spliceosome upon catalytic activation. Affinity-purified BactΔPrp2 complexes assembled on Atto647N-M3Act, carrying Yju2-EGFP (columns 1_–_3), were complemented with Prp2, Spp2, and ATP (columns 4_–_6), or with Prp2, Spp2, ATP, and Cwc25 (columns 7_–_9). After standard incubation, increasing concentrations of KCl were added.
FIGURE 7.
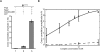
Catalytic activation by Prp2 creates a high-affinity binding site for Cwc25 in the spliceosome. (A) BactΔPrp2 complexes assembled on Atto647N-M3Act (column 1) were complemented on the amylose matrix with C-terminally labeled Cwc25-Alexa488 (column 2) or Cwc25-Alexa488 together with Prp2, Spp2 plus ATP (column 3). After incubation, the complexes were washed and eluted with maltose, and dcFCCS measurements were performed. (B) BactΔPrp2 complexes assembled on Atto647N-M3Act were directly eluted from the amylose matrix or activated on the column by Prp2 and Spp2 in the presence of ATP to generate B* before the elution. Increasing amounts of complexes were then supplemented with 0.3 nM Cwc25-Alexa488, and the resulting cross-correlation amplitude was measured. Cross-correlation amplitudes from two independent measurements were plotted against the respective complex concentration (Hill plot, semilogarithmic scale). Curve-fitting revealed a dissociation constant of Cwc25-Alexa488 to complex B* of _K_d = 0.03 nM. The lower curve was obtained under similar conditions but by adding complex BactΔPrp2 instead of B*.
FIGURE 8.
Schematic representations of the substantial remodeling the BactΔPrp2 spliceosome undergoes during the Prp2/ATP-mediated catalytic activation. Cwc24, Cwc27, the RES protein Bud13, Prp2, and Spp2 are released, while the U2 SF3a/b proteins are destabilized by Prp2/ATP, such that the BS adenosine becomes available for a nucleophile attack at the 5′ SS phosphodiester bond. At the same time, high-affinity binding sites are created for Yju2 and Cwc25, which are required to promote the first step of splicing.
Similar articles
- The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic RNA core of the spliceosome.
Wlodaver AM, Staley JP. Wlodaver AM, et al. RNA. 2014 Mar;20(3):282-94. doi: 10.1261/rna.042598.113. Epub 2014 Jan 17. RNA. 2014. PMID: 24442613 Free PMC article. - The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome.
Warkocki Z, Schneider C, Mozaffari-Jovin S, Schmitzová J, Höbartner C, Fabrizio P, Lührmann R. Warkocki Z, et al. Genes Dev. 2015 Jan 1;29(1):94-107. doi: 10.1101/gad.253070.114. Genes Dev. 2015. PMID: 25561498 Free PMC article. - DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps.
Tseng CK, Liu HL, Cheng SC. Tseng CK, et al. RNA. 2011 Jan;17(1):145-54. doi: 10.1261/rna.2459611. Epub 2010 Nov 22. RNA. 2011. PMID: 21098140 Free PMC article. - The RES complex is required for efficient transformation of the precatalytic B spliceosome into an activated Bact complex.
Bao P, Will CL, Urlaub H, Boon KL, Lührmann R. Bao P, et al. Genes Dev. 2017 Dec 1;31(23-24):2416-2429. doi: 10.1101/gad.308163.117. Epub 2018 Jan 12. Genes Dev. 2017. PMID: 29330354 Free PMC article. - Yeast Prp2 liberates the 5' splice site and the branch site adenosine for catalysis of pre-mRNA splicing.
Bao P, Höbartner C, Hartmuth K, Lührmann R. Bao P, et al. RNA. 2017 Dec;23(12):1770-1779. doi: 10.1261/rna.063115.117. Epub 2017 Sep 1. RNA. 2017. PMID: 28864812 Free PMC article.
Cited by
- The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction.
Liu HL, Cheng SC. Liu HL, et al. Mol Cell Biol. 2012 Dec;32(24):5056-66. doi: 10.1128/MCB.01109-12. Epub 2012 Oct 15. Mol Cell Biol. 2012. PMID: 23071087 Free PMC article. - Stoichiometries of U2AF35, U2AF65 and U2 snRNP reveal new early spliceosome assembly pathways.
Chen L, Weinmeister R, Kralovicova J, Eperon LP, Vorechovsky I, Hudson AJ, Eperon IC. Chen L, et al. Nucleic Acids Res. 2017 Feb 28;45(4):2051-2067. doi: 10.1093/nar/gkw860. Nucleic Acids Res. 2017. PMID: 27683217 Free PMC article. - The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction.
Fourmann JB, Dybkov O, Agafonov DE, Tauchert MJ, Urlaub H, Ficner R, Fabrizio P, Lührmann R. Fourmann JB, et al. Elife. 2016 Apr 26;5:e15564. doi: 10.7554/eLife.15564. Elife. 2016. PMID: 27115347 Free PMC article. - Biased Brownian ratcheting leads to pre-mRNA remodeling and capture prior to first-step splicing.
Krishnan R, Blanco MR, Kahlscheuer ML, Abelson J, Guthrie C, Walter NG. Krishnan R, et al. Nat Struct Mol Biol. 2013 Dec;20(12):1450-7. doi: 10.1038/nsmb.2704. Epub 2013 Nov 17. Nat Struct Mol Biol. 2013. PMID: 24240612 Free PMC article. - Dynamic Contacts of U2, RES, Cwc25, Prp8 and Prp45 Proteins with the Pre-mRNA Branch-Site and 3' Splice Site during Catalytic Activation and Step 1 Catalysis in Yeast Spliceosomes.
Schneider C, Agafonov DE, Schmitzová J, Hartmuth K, Fabrizio P, Lührmann R. Schneider C, et al. PLoS Genet. 2015 Sep 22;11(9):e1005539. doi: 10.1371/journal.pgen.1005539. eCollection 2015. PLoS Genet. 2015. PMID: 26393790 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases