Automatic design of synthetic gene circuits through mixed integer non-linear programming - PubMed (original) (raw)
Automatic design of synthetic gene circuits through mixed integer non-linear programming
Linh Huynh et al. PLoS One. 2012.
Abstract
Automatic design of synthetic gene circuits poses a significant challenge to synthetic biology, primarily due to the complexity of biological systems, and the lack of rigorous optimization methods that can cope with the combinatorial explosion as the number of biological parts increases. Current optimization methods for synthetic gene design rely on heuristic algorithms that are usually not deterministic, deliver sub-optimal solutions, and provide no guaranties on convergence or error bounds. Here, we introduce an optimization framework for the problem of part selection in synthetic gene circuits that is based on mixed integer non-linear programming (MINLP), which is a deterministic method that finds the globally optimal solution and guarantees convergence in finite time. Given a synthetic gene circuit, a library of characterized parts, and user-defined constraints, our method can find the optimal selection of parts that satisfy the constraints and best approximates the objective function given by the user. We evaluated the proposed method in the design of three synthetic circuits (a toggle switch, a transcriptional cascade, and a band detector), with both experimentally constructed and synthetic promoter libraries. Scalability and robustness analysis shows that the proposed framework scales well with the library size and the solution space. The work described here is a step towards a unifying, realistic framework for the automated design of biological circuits.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
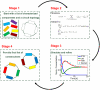
Figure 1. System overview of the proposed optimization framework.
The software requires access to a library of characterized parts (such as a subset of the parts available in Parts Registry) that will be used as fundamental blocks in the synthetic circuit. The user will have to supply a specific design (static connectivity), together with a set of constraints and a specific objective function to be optimized. The software will translate this system to a set of linear constraints that it will subsequently solve. The result of the optimization framework will be the set of parts that have to be used, and at what position. The system will have the ability to simulate the proposed design, and provide candidate synthetic circuits for experimental construction in the laboratory.
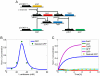
Figure 2. Band-pass filter design.
A) The system will only express the reporter when the concentration of the input signal (L-arabinose) is in a specific range. In this design, pCONST1 and pCONST2 are constitutive promoters, while pBAD1, pBAD2 and pLAC are the promoters where AraC and LacI bind, respectively. There are two coding regions of TetR which are put on the downstream of promoters pBAD2 and pLAC. In the absence of L-arabinose, AraC activates TetR production by de-repressing the pLAC promoter. In high L-arabinose concentrations, TetR is again produced through the de-repression of the pBAD2 promoter. In significant, but not high inducer concentrations, however, none of the pathways are active enough, which in turn results in lower TetR levels and subsequent expression of the reporter GFP output. B) Reporter protein concentration (output) versus L-arabinose levels (input). The output of the synthetic circuit becomes high only at moderate values of L-arabinose. Green circles denote desired values (fluorescence measurements) that act as input to our optimization platform. C) Temporal expression profile of the band-pass filter. Temporal profile of the resulting optimal synthetic gene circuit, for a L-arabinose level of 30 mM. The GFP concentration of the optimization-derived circuit (cyan solid line) matches well the desired input values (yellow solid dots).
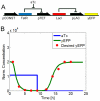
Figure 3. Transcriptional cascade design.
A) The system is controlled by the inducer aTc which can bind to TetR and reduce the concentration of free TetR molecules. This concentration change will be propagated through the cascade to the change of the reporter yEFP. B) Temporal profile of a cascade design: The desired output (yEFP, red dots) and the actual output (green line) of the optimal synthetic gene circuit are showed. The temporal profile was split into two phases, based on changes in the inducer concentration. In the first phase (0 h–9 h), 2.16  aTc (blue line) is added and in the second phase (9 h–24 h) aTc is washed out (setting as the experiment in [24]).
aTc (blue line) is added and in the second phase (9 h–24 h) aTc is washed out (setting as the experiment in [24]).
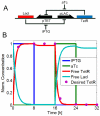
Figure 4. Toggle switch design, where two genes (LacI and TetR) negatively regulate each other.
A) The system is externally controlled through the addition of two inducers, IPTG and aTc, which bind to the repressors and decrease their regulatory potential. B) Expression profile of the resulting synthetic circuit: The desired profile (input, depicted with purple dots) and actual profile (red line) for the TetR protein is shown. The temporal profile was split into four phases, based on changes in the inducer concentrations. Phase 1: IPTG high, aTc low; Phase 2: IPTG low, aTc low; Phase 3: IPTG low, aTc high; Phase 4: IPTG low, aTc low.
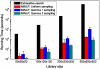
Figure 5. Scalability and sensitivity of the running time of the MINLP approach on different inputs.
A comparison of the running time (seconds) of the MINLP approach on different input sizes and different data input distribution for the steady state cascade design problem (5 synthetic datasets per case). Parameter values are generated by sampling within the parameter range that has been experimentally measured –. Samples are distributed either uniformly (uniform sampling), following a gamma distribution with the mean set on the optimal set parameter values (Gamma 1 sampling), or away from that mean (Gamma 2 sampling). The running time of exhaustive search method (ES) is estimated for the last library size.
Similar articles
- Optimal part and module selection for synthetic gene circuit design automation.
Huynh L, Tagkopoulos I. Huynh L, et al. ACS Synth Biol. 2014 Aug 15;3(8):556-64. doi: 10.1021/sb400139h. Epub 2014 Feb 27. ACS Synth Biol. 2014. PMID: 24933033 - Automated Biocircuit Design with SYNBADm.
Otero-Muras I, Banga JR. Otero-Muras I, et al. Methods Mol Biol. 2021;2229:119-136. doi: 10.1007/978-1-0716-1032-9_4. Methods Mol Biol. 2021. PMID: 33405218 - OptCircuit: an optimization based method for computational design of genetic circuits.
Dasika MS, Maranas CD. Dasika MS, et al. BMC Syst Biol. 2008 Mar 3;2:24. doi: 10.1186/1752-0509-2-24. BMC Syst Biol. 2008. PMID: 18315885 Free PMC article. - [Machine learning-aided design of synthetic biological parts and circuits].
Mao R, Wang B. Mao R, et al. Sheng Wu Gong Cheng Xue Bao. 2025 Mar 25;41(3):1023-1051. doi: 10.13345/j.cjb.240605. Sheng Wu Gong Cheng Xue Bao. 2025. PMID: 40170311 Review. Chinese. - Engineering synthetic regulatory circuits in plants.
Kassaw TK, Donayre-Torres AJ, Antunes MS, Morey KJ, Medford JI. Kassaw TK, et al. Plant Sci. 2018 Aug;273:13-22. doi: 10.1016/j.plantsci.2018.04.005. Epub 2018 Apr 11. Plant Sci. 2018. PMID: 29907304 Review.
Cited by
- Protein-based bandpass filters for controlling cellular signaling with chemical inputs.
Shui S, Scheller L, Correia BE. Shui S, et al. Nat Chem Biol. 2024 May;20(5):586-593. doi: 10.1038/s41589-023-01463-7. Epub 2023 Nov 13. Nat Chem Biol. 2024. PMID: 37957273 Free PMC article. - Multicriteria global optimization for biocircuit design.
Otero-Muras I, Banga JR. Otero-Muras I, et al. BMC Syst Biol. 2014 Sep 24;8:113. doi: 10.1186/s12918-014-0113-3. BMC Syst Biol. 2014. PMID: 25248337 Free PMC article. - How to make a synthetic multicellular computer.
Macia J, Sole R. Macia J, et al. PLoS One. 2014 Feb 19;9(2):e81248. doi: 10.1371/journal.pone.0081248. eCollection 2014. PLoS One. 2014. PMID: 24586222 Free PMC article. - Designing synthetic networks in silico: a generalised evolutionary algorithm approach.
Smith RW, van Sluijs B, Fleck C. Smith RW, et al. BMC Syst Biol. 2017 Dec 2;11(1):118. doi: 10.1186/s12918-017-0499-9. BMC Syst Biol. 2017. PMID: 29197394 Free PMC article. - Synthetic biology outside the cell: linking computational tools to cell-free systems.
Lewis DD, Villarreal FD, Wu F, Tan C. Lewis DD, et al. Front Bioeng Biotechnol. 2014 Dec 9;2:66. doi: 10.3389/fbioe.2014.00066. eCollection 2014. Front Bioeng Biotechnol. 2014. PMID: 25538941 Free PMC article. Review.
References
- Mukhopadhyay A, Redding A, Rutherford B, Keasling J. Importance of systems biology in engineering microbes for biofuel production. Current opinion in biotechnology. 2008;19:228–234. - PubMed
- Densmore D, Kittleson J, Bilitchenko L, Liu A, Anderson J. Rule based constraints for the construction of genetic devices. 2010. pp. 557–560. In: Circuits and Systems (ISCAS), Proceedings of 2010 IEEE International Symposium on. IEEE.