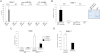A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy - PubMed (original) (raw)
A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy
Daphne S Cabianca et al. Cell. 2012.
Abstract
Repetitive sequences account for more than 50% of the human genome. Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal-dominant disease associated with reduction in the copy number of the D4Z4 repeat mapping to 4q35. By an unknown mechanism, D4Z4 deletion causes an epigenetic switch leading to de-repression of 4q35 genes. Here we show that the Polycomb group of epigenetic repressors targets D4Z4 in healthy subjects and that D4Z4 deletion is associated with reduced Polycomb silencing in FSHD patients. We identify DBE-T, a chromatin-associated noncoding RNA produced selectively in FSHD patients that coordinates de-repression of 4q35 genes. DBE-T recruits the Trithorax group protein Ash1L to the FSHD locus, driving histone H3 lysine 36 dimethylation, chromatin remodeling, and 4q35 gene transcription. This study provides insights into the biological function of repetitive sequences in regulating gene expression and shows how mutations of such elements can influence the progression of a human genetic disease.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
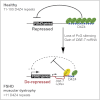
Graphical abstract
Figure 1
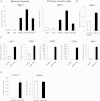
D4Z4 Recruits PcG Complexes to Repress 4q35 Genes (A–C) ChIP for EZH2, H3K27me3, and IgG on primary muscle cells from healthy donors and FSHD patients. A scheme of the FSHD locus primers used for the analysis (A). ChIP analyzed by qPCR for p13E-11, nondeleted element (NDE), and different regions of the D4Z4 repeat. Results are expressed as relative enrichment to input normalized for repeat amount (B), or percentage of total H3 (C). The mean of the signals obtained from four healthy samples or four FSHD patients is shown. The error bars represent SEM. Asterisks indicate statistical significance (p value) as evaluated by two-way repeated-measured ANOVA, respectively p = 0.0051 (B) and p = 0.0497 (C). (D–F) CHO cells stably transfected with human 4q35 BACs either containing (CH16-291A23) or lacking (RP11-462G22) 4q35 D4Z4 repeats (schematized in D), were analyzed by ChIP for core components of PRC1 and H2Aub1 (E) and PRC2 and H3K27me3 (F). ChIP was analyzed by qPCR with primers for a region common to both BACs (see scheme D). Results are expressed as percentage of input (PcG proteins), percentage of total H3 (H3K27me3), or percentage of H2A, normalized to IgM (H2Aub1). The error bars represent SEM. (G–J) Chr4/CHO cells stably expressing control shRNA or sh_Suz12_. Knockdown was evaluated by qRT-PCR (G) and immunoblotting (H). Expression of Hoxd11, Gapdh (I) and 4q35 genes ANT1, FRG1, FRG2, and DUX4 was analyzed by qRT-PCR (J) and expressed over β-actin. The error bars represent SEM. See also Figures S1 and S2.
Figure 2
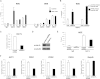
The DBE Region Is Transcribed Selectively in FSHD Patients or FSHD-Like Conditions (A) qRT-PCR analysis of DBE-T in muscle biopsies (left) and primary muscle cells (right) from healthy donors and FSHD patients. Results are expressed over GAPDH. (B) qRT-PCR analysis of DBE-T in control or Suz12 knockdown chr4/CHO cells. Results are expressed over β-actin. (C) qRT-PCR analysis of 4q35 genes and DBE-T in chr4/CHO cells in the repressed (control) or de-repressed (AZA+TSA) states. Results are expressed over β-actin. (D) qRT-PCR analysis of Hoxd11 and Gapdh in chr4/CHO cells in the repressed (control) or de-repressed (AZA+TSA) states. Results are expressed over β-actin. The error bars represent SEM. See also Figure S3.
Figure 3
DBE-T Is Required for De-Repression and Topological Reorganization of the 4q35 Region (A) Control or DBE-T chr4/CHO knockdown cells were treated with AZA+TSA. DBE-T knockdown, 4q35 genes and Gapdh expression were evaluated by qRT-PCR. Results are expressed over β-actin. The error bars represent SEM. (B) Schematic representation of the region analyzed by 3C. The D1 3C primer, located near DBE, was used as bait and primers spanning the FRG1 genomic locus were tested for interaction. 3C results show the relative frequency of interaction between DBE and FRG1 locus in control and AZA+TSA treated chr4/CHO cells expressing a nonsilencing shRNA (C left) or an shRNA specific for DBE-T (sh_DBE-T_) (C right). The error bars represent SEM. See also Figures S4 and S5.
Figure 4
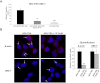
DBE-T Is Nuclear, Chromatin-Associated, and Localized to the FSHD Locus (A) Total RNA from AZA+TSA treated chr4/CHO cells was separated into cytoplasmic, nuclear-soluble, and chromatin-bound fractions. The relative abundance of DBE-T in the different fractions was measured by qRT-PCR. As control, Gapdh and Xist were analyzed. The error bars represent SEM. (B) Schematic representation of the location of the DBE-T and D4Z4 probes. (C) Following AZA+TSA treatment, chr4/CHO cells were analyzed by RNA/DNA FISH. A single Z stack acquired with an Olympus IX70 DeltaVision RT Deconvolution System microscope is shown. Signals colocalization was detected in 98.8% of the double positive cells (n = 80) deriving from 3 independent experiments. DBE-T is visualized in green, and the D4Z4 DNA is in red. DAPI is in blue. See also Figures S6 and S7.
Figure 5
The TrxG Protein Ash1L Is Recruited to the FSHD Locus and De-Represses 4q35 Genes (A) ChIP for Ash1L, Chd7, Mll1, and IgG on chr4/CHO cells in the repressed (control) or de-repressed (AZA+TSA) states. ChIP was analyzed by qPCR with primers for NDE and DBE and expressed as percentage of input. The error bars represent SEM. (B) ChIP for ASH1L in control and FSHD primary muscle cells. ChIP was analyzed by qPCR with primers for NDE and expressed as percentage of input normalized to IgG. The error bars represent SEM. (C–F) Chr4/CHO cells stably expressing control shRNA or sh_Ash1L_. Knockdown after AZA+TSA treatment evaluated by qRT-PCR (C), immunoblotting (D), and ChIP on NDE (E). Expression of 4q35 genes and Gapdh, as control, was analyzed by qRT-PCR (F). Results are expressed over β-actin. The error bars represent SEM.
Figure 6
DBE-T Directly Binds the TrxG Protein Ash1L and Recruits It to the FSHD Locus (A) RNA immunoprecipitation (IP) following UV crosslinking for Ash1L or IgG on AZA+TSA treated chr4/CHO cells. DBE-T or, as control, pre-miR19A and U1 snRNA enrichments were measured by qRT-PCR. The error bars represent SEM. (B) In vitro RNA-GST pull-down assay showing the interaction between recombinant GST-fused Ash1L SET domain or GST and in vitro transcribed DBE-T. On the right, Coomassie staining of purified recombinant proteins. After RNA recovery, samples were analyzed by qRT-PCR. The error bars represent SEM. (C) Following AZA+TSA treatment, chr4/CHO cells stably expressing a nonsilencing control shRNA or sh_DBE-T_ were analyzed by ChIP for Ash1L or IgG. Enrichment for NDE was analyzed by qPCR and displayed as enrichment relative to input. The error bars represent SEM. (D) Upon AZA+TSA treatment, control shRNA, and sh_Ash1L_ cells were collected to analyze DBE-T expression by qRT-PCR. Results are expressed over β-actin. The error bars represent SEM. See also Figure S8.
Figure 7
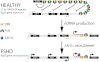
Model for _DBE-T_-Mediated 4q35 Gene De-Repression in FSHD Normal individuals carry multiple D4Z4 copies that are extensively bound by PcG proteins, promoting the maintenance of repressed chromatin at 4q35. In FSHD patients, D4Z4 deletion leads to insufficient binding of PcG, causing the production of DBE-T. DBE-T recruits the TrxG protein ASH1L and promotes a topological reorganization of the FSHD locus leading to de-repression of 4q35 genes.
Figure S1
The FSHD Locus Displays Similarities with Drosophila Polycomb Response Elements, Related to Figure 1 Occurrence in the FSHD locus of DNA motifs found in Drosophilia PREs/TREs: PRE consensus; Pleiohomeotic/Pleiohomeotic-like (Pho/PhoL) core consensus; GAGAG motif (GAF). A scheme of the FSHD locus is also shown. DNA motifs are highlighted on the DNA sequence in a portion of D4Z4 repeat (GenBank: AF117653.2; 6661-7980 bp).
Figure S2
PcG Core Components and the Associated Histone Marks Are Enriched at the FSHD Locus, Related to Figure 1 ChIP-qPCR experiments with antibodies specific for EZH2 (A) and its associated histone mark, H3K27me3 (B), in primary muscle cells from 4 healthy donors and 4 FSHD patients. ChIP was analyzed by qPCR with primers specific for the promoters of 4q35 genes ANT1, FRG1 and FRG2. NDE is shown as comparison of the enrichment in the FSHD locus. Results are expressed as percentage of input (EZH2), or percentage of total H3 (H3K27me3). The mean of the signals obtained from 4 healthy samples or 4 FSHD patients is shown. The error bars represent SEM. (C) Scheme of the FSHD locus. The location of the primers used for ChIP-qPCR is shown. (D-G) ChIP-qPCR experiments with antibodies specific for the core components of PRC1 and H2Aub1 (D), PRC2 and H3K27me3 (E), and Jarid2 and c-Krox/Th-POK (F) and macroH2A (G) in human chr4/CHO. Hoxd11 is shown as positive control. ChIP material was analyzed by qPCR with primers for different regions of the D4Z4 repeat and for NDE (see scheme C). Results are expressed as percentage of input (PcG proteins and macroH2A), percentage of the ChIP signal obtained with anti-H3 antibodies (H3K27me3) or percentage of the ChIP signal obtained with anti-H2A antibodies and normalized for IgM (H2Aub1). The error bars represent SEM.
Figure S3
AZA and TSA Treatments Generate Specific Effects, Related to Figure 2 (A) Relative qPCR analysis showing that the amount of D4Z4 repeats is unaffected by AZA plus TSA treatment. The error bars represent SEM. (B-C) ChIP-qPCR experiments with antibodies specific for Ezh2 (B) and its associated histone mark, H3K27me3 (C), in chr4/CHO cells treated with AZA. Ezh2 and H3K27me3 signals are not reduced by AZA treatment. ChIP was analyzed by qPCR with primers specific for NDE. Results are expressed as percentage of input (Ezh2), or percentage of total H3 (H3K27me3). The error bars represent SEM.
Figure S4
DBE-T shRNA Does Not Cause Direct Transcriptional Gene Silencing, Related to Figure 3 Following AZA+TSA treatment, human chr4/CHO hybrid cells stably expressing a nonsilencing control shRNA or sh_DBE-T_ were collected to extract chromatin. ChIP for Ago1 or IgG, as control was performed. Enrichment at the β-actin promoter, NDE and DBE regions was analyzed by qPCR. Results are displayed as percentage of input. The error bars represent SEM.
Figure S5
DBE-T does not function in trans, related to Figure 3 DBE-T was ectopically overexpressed in human chr4/CHO cells and expression of 4q35 genes and Gapdh, as control, was analyzed by qRT-PCR. Results are shown as expression over β-actin. The error bars represent SEM.
Figure S6
DBE-T Is a Mature RNA, Related to Figure 4 (A) Human chr4/CHO and CHO cells were collected in the de-repressed state (AZA+TSA). A set of samples were treated with RNase A+T1. RNA FISH quantification of DBE-T positive nuclei was performed in all conditions and it was normalized to signals in human chr4/CHO cells treated with AZA+TSA. The error bars represent SD. (B) AZA+TSA human chr4/CHO cells were treated (Right panels) or untreated (Left panels) with Actinomycin D at 5 μg/ml for 15′. Hybridization to β-actin (Top panels) or DBE-T (Bottom panels) RNAs was performed. The arrows indicate dots of nuclear RNAs. See Statistical Test section for statistical analysis. The error bars represent SD. DBE-T and β-actin RNAs are in red. DAPI is in blue. The images correspond to a single Z stack acquired with an Olympus IX70 DeltaVision RT Deconvolution System microscope. (A and B) The asterisks indicate statistically significant differences. A Two-tailed, paired, t test was applied. For DBE-T: human chr4/CHO AZA+TSA n = 4; human chr4/CHO AZA+TSA + Actinomycin D n = 4, p = 0.0914; human Chr4/CHO AZA+TSA + RNase A+T1 n = 4, p = 0.0056; CHO AZA+TSA n = 4, p = 0.0006. For β-actin: human chr4/CHO AZA+TSA n = 4; human chr4/CHO AZA+TSA + Actinomycin D n = 4, p = 0.0012.
Figure S7
DBE-T Is a Long ncRNA Whose Transcription Starts outside the D4Z4 Repeat Array and Encompasses the NDE Region, Related to Figure 4 (A) Schematic representation of the regions analyzed. The asterisk corresponds to the Transcriptional Start Site (TSS) mapped by 5′ RACE. The position indicated is referred to AF117653. The arrows represent the primers used in panel C to amplify a single NDE/DBE-T transcript. Lines and numbers positioned in the lower part of the scheme correspond to the overlapping regions amplified in panel D. (B) Northern blot assays performed on PolyA+ RNA extracted from human chr4/CHO cells untreated (Control) or treated with AZA+TSA (AZA+TSA). Hybridizations with probes mapping to NDE, DBE, DUX4 and Gapdh, as loading control, are shown. (C) RT-PCR to amplify a single transcript from NDE to DBE was performed on RNA extracted from CHO cells transfected with a construct carrying the entire AF117653 sequence (pGEM42, derived from an FSHD patient, containing the 4q35 region ranging from upstream p13E-11, two D4Z4 repeats and the distal region, Kowaljow et al., 2007) or with the empty vector as control. As positive control, the pGEM42 plasmid DNA was PCR amplified; RT- and no template reactions were performed as negative controls. (D) The NDE-DBE region was divided in eight overlapping regions (see panel A) that were amplified by RT-PCR in RNA samples extracted from human chr4/CHO cells untreated (Control) or treated with AZA+TSA (AZA+TSA). As loading control, Gapdh was amplified. (E) NDE transcription was evaluated in the repressed (control) and de-repressed (AZA+TSA) states by qRT-PCR. Results are shown as expression over β-actin. The error bars represent SEM. (F) The relative enrichment of the NDE transcript in the indicated subcellular fractions as measured by qRT-PCR is shown. The error bars represent SEM. (G) Analysis of NDE expression by qRT-PCR in control and FSHD primary muscle cells. Results are shown as expression over GAPDH. The error bars represent SEM. (H) Expression of 4q35 genes and Gapdh, as control, in human chr4/CHO cells knockdown for NDE transcript (G left) in the de-repressed state (AZA+TSA). Results are shown as expression over β-actin. The error bars represent SEM. (I) The NDE transcript is downregulated upon DBE-T knockdown and DBE-T is downregulated upon NDE transcript knockdown. Results are shown as expression over β-actin. The error bars represent SEM.
Figure S8
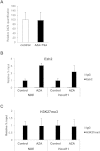
Ash1L Dimethylates Lysine 36 on Histone H3 at the FSHD Locus, Related to Figure 6 (A) ChIP-qPCR for enrichment on total H3 of H3K4me3, H3K36me2 and IgG, as control, performed on human chr4/CHO cells in the repressed (control) or in the de-repressed (AZA+TSA) state. ChIP material was analyzed by qPCR with primers specific for the NDE region. The error bars represent SEM. (B) ChIP-qPCR for enrichment on total H3 of H3K4me3, H3K36me2 and IgG, as control, performed in control shRNA or sh_Ash1L_ expressing cells in the de-repressed state (AZA+TSA). ChIP material was analyzed by qPCR with primers specific for the NDE region. The error bars represent SEM. (C) ChIP-qPCR for enrichment on total H3 of H3K36me2 and IgG, as control, in control shRNA or sh_DBE-T_ expressing cells in the de-repressed state (AZA+TSA). ChIP material was analyzed by qPCR with primers specific for the NDE region. The error bars represent SEM. (D) ChIP for Ash1L and IgG, as control, on human chr4/CHO cells in the repressed (control) or de-repressed (AZA+TSA) state. Analysis was performed by qPCR with primers specific for ANT1, FRG1, FRG2 promoters and NDE. Results are expressed as relative percentage of input. The error bars represent SEM. (E) ChIP-qPCR for enrichment on total H3 of H3K36me2 and IgG, as control, performed on human chr4/CHO cells in the repressed (control) or in the de-repressed (AZA+TSA) state. Analysis was performed by qPCR with primers specific for ANT1, FRG1, FRG2 promoters and NDE. Results are expressed as relative percentage of H3. The error bars represent SEM.
Similar articles
- The activatory long non-coding RNA DBE-T reveals the epigenetic etiology of facioscapulohumeral muscular dystrophy.
Vizoso M, Esteller M. Vizoso M, et al. Cell Res. 2012 Oct;22(10):1413-5. doi: 10.1038/cr.2012.93. Epub 2012 Jun 19. Cell Res. 2012. PMID: 22710800 Free PMC article. - Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation.
Bodega B, Ramirez GD, Grasser F, Cheli S, Brunelli S, Mora M, Meneveri R, Marozzi A, Mueller S, Battaglioli E, Ginelli E. Bodega B, et al. BMC Biol. 2009 Jul 16;7:41. doi: 10.1186/1741-7007-7-41. BMC Biol. 2009. PMID: 19607661 Free PMC article. - Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD).
Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, Kimonis VE, Balog J, Frants RR, Ball AR Jr, Lock LF, Donovan PJ, van der Maarel SM, Yokomori K. Zeng W, et al. PLoS Genet. 2009 Jul;5(7):e1000559. doi: 10.1371/journal.pgen.1000559. Epub 2009 Jul 10. PLoS Genet. 2009. PMID: 19593370 Free PMC article. - In junk we trust: repetitive DNA, epigenetics and facioscapulohumeral muscular dystrophy.
Neguembor MV, Gabellini D. Neguembor MV, et al. Epigenomics. 2010 Apr;2(2):271-87. doi: 10.2217/epi.10.8. Epigenomics. 2010. PMID: 22121874 Review. - A complex interplay of genetic and epigenetic events leads to abnormal expression of the DUX4 gene in facioscapulohumeral muscular dystrophy.
Gatica LV, Rosa AL. Gatica LV, et al. Neuromuscul Disord. 2016 Dec;26(12):844-852. doi: 10.1016/j.nmd.2016.09.015. Epub 2016 Sep 19. Neuromuscul Disord. 2016. PMID: 27816329 Review.
Cited by
- DUX4 differentially regulates transcriptomes of human rhabdomyosarcoma and mouse C2C12 cells.
Sharma V, Harafuji N, Belayew A, Chen YW. Sharma V, et al. PLoS One. 2013 May 22;8(5):e64691. doi: 10.1371/journal.pone.0064691. Print 2013. PLoS One. 2013. PMID: 23717650 Free PMC article. - The activatory long non-coding RNA DBE-T reveals the epigenetic etiology of facioscapulohumeral muscular dystrophy.
Vizoso M, Esteller M. Vizoso M, et al. Cell Res. 2012 Oct;22(10):1413-5. doi: 10.1038/cr.2012.93. Epub 2012 Jun 19. Cell Res. 2012. PMID: 22710800 Free PMC article. - Genetic "lnc"-age of noncoding RNAs to human disease.
Troy A, Sharpless NE. Troy A, et al. J Clin Invest. 2012 Nov;122(11):3837-40. doi: 10.1172/JCI66645. Epub 2012 Oct 24. J Clin Invest. 2012. PMID: 23093789 Free PMC article. - HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition.
Bonomi S, di Matteo A, Buratti E, Cabianca DS, Baralle FE, Ghigna C, Biamonti G. Bonomi S, et al. Nucleic Acids Res. 2013 Oct;41(18):8665-79. doi: 10.1093/nar/gkt579. Epub 2013 Jul 17. Nucleic Acids Res. 2013. PMID: 23863836 Free PMC article. - DICER/AGO-dependent epigenetic silencing of D4Z4 repeats enhanced by exogenous siRNA suggests mechanisms and therapies for FSHD.
Lim JW, Snider L, Yao Z, Tawil R, Van Der Maarel SM, Rigo F, Bennett CF, Filippova GN, Tapscott SJ. Lim JW, et al. Hum Mol Genet. 2015 Sep 1;24(17):4817-28. doi: 10.1093/hmg/ddv206. Epub 2015 Jun 3. Hum Mol Genet. 2015. PMID: 26041815 Free PMC article.
References
- Bakker E., Wijmenga C., Vossen R.H., Padberg G.W., Hewitt J., van der Wielen M., Rasmussen K., Frants R.R. The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve. 1995;2:S39–S44. - PubMed
- Beisel C., Paro R. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 2011;12:123–135. - PubMed
Supplemental References
- Ohtsubo, M., Yasunaga, S., Ohno, Y., Tsumura, M., Okada, S., Ishikawa, N., Shirao, K., Kikuchi, A., Nishitani, H., Kobayashi, M., and Takihara, Y. (2008). Polycomb-group complex 1 acts as an E3 ubiquitin ligase for Geminin to sustain hematopoietic stem cell activity. Proc. Natl. Acad. Sci. USA 105, 10396–10401. - PMC - PubMed
- Sambrook, J., and Russel, D.W. (2001). Molecular cloning: a laboratory manual (Cold Spring Harbor, NY: Cold Harbor Spring Laboratory Press).
- Stock, J.K., Giadrossi, S., Casanova, M., Brookes, E., Vidal, M., Koseki, H., Brockdorff, N., Fisher, A.G., and Pombo, A. (2007). Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 9, 1428–1435. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials