A self-produced trigger for biofilm disassembly that targets exopolysaccharide - PubMed (original) (raw)
A self-produced trigger for biofilm disassembly that targets exopolysaccharide
Ilana Kolodkin-Gal et al. Cell. 2012.
Retraction in
- Retraction Notice to: A Self-Produced Trigger for Biofilm Disassembly that Targets Exopolysaccharide.
[No authors listed] [No authors listed] Cell. 2015 May 7;161(4):946. doi: 10.1016/j.cell.2015.04.039. Cell. 2015. PMID: 25984577 Free PMC article. No abstract available.
Abstract
Biofilms are structured communities of bacteria that are held together by an extracellular matrix consisting of protein and exopolysaccharide. Biofilms often have a limited lifespan, disassembling as nutrients become exhausted and waste products accumulate. D-amino acids were previously identified as a self-produced factor that mediates biofilm disassembly by causing the release of the protein component of the matrix in Bacillus subtilis. Here we report that B. subtilis produces an additional biofilm-disassembly factor, norspermidine. Dynamic light scattering and scanning electron microscopy experiments indicated that norspermidine interacts directly and specifically with exopolysaccharide. D-amino acids and norspermidine acted together to break down existing biofilms and mutants blocked in the production of both factors formed long-lived biofilms. Norspermidine, but not closely related polyamines, prevented biofilm formation by B. subtilis, Escherichia coli, and Staphylococcus aureus.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
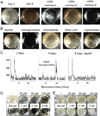
Figure 1. Identification of norspermidine in conditioned medium from B. subtilis and its effect on pellicle formation
Panel A. Biofilm-inhibiting factors in conditioned medium
. B. subtilis strain NCBI3610 was grown at 22 °C in 12-well plates in liquid biofilm-inducing medium for 3 or 8 days. Conditioned medium (500 ml) from an 8-day-old culture was concentrated on the C-18 column and eluted step-wise with methanol. Shown is the result of growing cells in fresh medium to which had been added 20 µl of the 25%, 35% or 40% methanol eluates.
Panel B. Norspermidine inhibits biofilm formation.
Cells of NCBI3610 were grown in fresh medium containing PBS buffer (control), norspermidine (100 µM), morpholine (100 µM) HPLC-purified fatty acid (~100 µM), or spermidine (100 µM). Brighter images of the norspermidine-treated cell revealed cells near the bottom of the well.
Panel C. Detection of norspermidine.
Pellicles were collected from 3- and 8-day-old cultures (100 ml) of the wild type (NCBI3610) and from an 8-day-old culture (100 ml) of a gabT mutant (IKG623). After mild sonication of the pellicles, cells were separated from extracellular material. (Other experiments showed that norspermidine is largely found in pellicles.) Norspermidine in the extracellular material was derivatized with Fmoc-Cl and the resulting Fmoc-norspermidine was detected using an Agilent LC/MS system. Fmoc-norspermidine was detectable in the old pellicle from wild type cells but not in the young or mutant pellicles. See also Figure S1, Panel B.
Panels D and E. Quanitification of the biofilm-inhibiting activity of norspermidine and spermidine).
Pellicle formation of strain NCBI3610 was tested in the presence of the indicated concentrations of norspermidine (D) or spermidine (E). See also Figure S1, Panel A.
Figure 2. Norspermidine acts together with D-amino acids
Panel A. Pellicle longevity.
Shown are 7 day-old cultures of the wild type (WT), a mutant (Δ_gbaT_) blocked in norspermidine production (IKG623), a double mutant (Δ_ylmE_ Δ_racX_) blocked in D-amino acid production (IKG55) and a triple mutant (Δ_gbaT_ Δ_ylmE_ Δ_racX_) blocked in the production of both (IKG625). See also Figure S2, Panel A.
Panel B. Preventing biofilm formation
. Cells were grown for 3 days in medium containing as indicated D-tyrosine (D-Tyr), norspermidine, a mixture of D-tyrosine, D-methionine, D-leucine and D-tryptophan (D-aa) and the indicated combinations of amino acids and norspermidine at the indicated concentrations.
Panel C. Qunatifying pellicle breakdown.
On the surface of 3 day-old pellicles were placed droplets (50 µl) containing buffer (PBS), a mixture of D-tyrosine, D-methionine, D-leucine and D-tryptophan each at final concentration of 12.5 µM, norspermidine at a final concentration of 50 µM, or, as in panel C, a combination of D-amino acids each at a concentration of only 2.5 µM and norspermidine at a concentration of 10 µM. After incubation for the indicated times, pellicle material and the medium were separated and each brought to a volume of 3 ml. After mild sonication, the OD600 was determined for each sample. The % of disassembly represents the OD600 of the medium as a percent of the sum of the OD600 of the medium and the OD600 of the pellicle.
Figure 3. Norspermidine disrupts exopolysaccharide
Shown are phase contrast and fluorescence images of cells of the wild-type (WT; NCBI3610) harvested from pellicles grown in the presence or absence (untreated) of norspermidine (25 µM) or a high concentration of spermidine (1 mM). The cells were washed in PBS and stained for exopolysaccharide with a conjugate of concanavalin A with Texas-Red. See also Figures S2, Panel B, and Figure S3
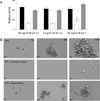
Figure 4. Norspermidine interacts with exopolysaccharide polymers
Panel A. Dynamic light scattering
. Listed are the average hydrodynamic radii of the exopolysaccharide as measured by dynamic light scattering. Exopolysaccharide was purified from pellicles. Light scattering was measured for exopolysaccharide alone as well as for exopolysaccharide that had been mixed with 0.75 mM norspermidine or with 0.75 mM spermidine. Shown are the results obtained in the absence of polyamine (black), in the presence of norspermidine (white), and in the presence of spermidine (grey) with exopolysaccharide at the indicated concentrations and pH. Error bars represent the standard deviation of polymer radii among the polymers in a single sample.
Panel B. Scanning Electron Microscopy
Purified exopolysaccharide was dissolved in double distilled water at a final concentration of 10 mg/ml and mixed with either norspermidine or spermidine (0.75 mM final concentration). Samples were prepared as described in Experimental Procedures. Shown are three different magnifications of representative fields showing exopolysaccharide alone (EPS) and exopolysaccharide that had been mixed with norspermidine (EPS + norspermidine) or with spermidine (EPS + spermidine). See Figure S4 for controls showing little effect on growth or eps transcription.
Figure 5. Structure activity relationship study of norspermidine
Panel A shows compounds tested for biofilm-inhibiting activity.
Panel B shows the effect of the numbered compounds on pellicle formation by B. subtilis (NCBI3610).
The compounds were tested at 200 µM.
Panels C and D show the results of modeling the interaction of norspermidine and spermidine with an acidic exopolysaccharide.
Norspermidine binds via salt bridges between amino and carboxyl groups (dotted lines) in a clamp-like mode across the exopolysaccharide secondary structure of a disaccharide repeat. [α(1,6)Glc-β(1,3)GlcA]n. Whereas norspermidine aligns well with the repeat, the spacing of amino groups of spermidine does not match the symmetric pattern of anionic side groups, implying weaker affinity. See supporting information for modelling of plausible interactions with other charged and non-charged polysaccharide structures. For tests of additional molecules see Figure S5 and Table S2.
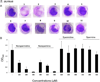
Figure 6. Norspermidine inhibits biofilm formation by S. aureus
Panel A shows the effect of the numbered compounds displayed in Figure 5A on the formation of submerged biofilms by S. aureus strain SCO1. The compounds were tested at 500 µM. Biofilm formation was visualized by crystal violet staining of submerged biofilms. Panel B shows quantification of the effects of norspermidine, norspermine, spermine and spermidine as measured by crystal violet staining (see Experimental procedures). See also Figure S6.
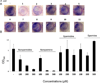
Figure 7. Norspermidine inhibits biofilm formation by E. coli
Panel A shows the effect of the numbered compounds shown in Figure 5A on submerged biofilm formation by E. coli strain MC4100. The compounds were tested at 500 µM. Biofilm formation was visualized by crystal violet staining of submerged biofilms. Panel B shows quantification of the effects of norspermidine, norspermine, spermine and spermidine as measured by crystal violet staining (see Experimental procedures). See also Figure S6.
Similar articles
- Norspermidine is not a self-produced trigger for biofilm disassembly.
Hobley L, Kim SH, Maezato Y, Wyllie S, Fairlamb AH, Stanley-Wall NR, Michael AJ. Hobley L, et al. Cell. 2014 Feb 13;156(4):844-54. doi: 10.1016/j.cell.2014.01.012. Cell. 2014. PMID: 24529384 Free PMC article. - Spermidine promotes Bacillus subtilis biofilm formation by activating expression of the matrix regulator slrR.
Hobley L, Li B, Wood JL, Kim SH, Naidoo J, Ferreira AS, Khomutov M, Khomutov A, Stanley-Wall NR, Michael AJ. Hobley L, et al. J Biol Chem. 2017 Jul 21;292(29):12041-12053. doi: 10.1074/jbc.M117.789644. Epub 2017 May 25. J Biol Chem. 2017. PMID: 28546427 Free PMC article. - Inhibitory effects of D-amino acids on Staphylococcus aureus biofilm development.
Hochbaum AI, Kolodkin-Gal I, Foulston L, Kolter R, Aizenberg J, Losick R. Hochbaum AI, et al. J Bacteriol. 2011 Oct;193(20):5616-22. doi: 10.1128/JB.05534-11. Epub 2011 Aug 19. J Bacteriol. 2011. PMID: 21856845 Free PMC article. - Biofilms.
López D, Vlamakis H, Kolter R. López D, et al. Cold Spring Harb Perspect Biol. 2010 Jul;2(7):a000398. doi: 10.1101/cshperspect.a000398. Epub 2010 Jun 2. Cold Spring Harb Perspect Biol. 2010. PMID: 20519345 Free PMC article. Review. - Sticking together: building a biofilm the Bacillus subtilis way.
Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Vlamakis H, et al. Nat Rev Microbiol. 2013 Mar;11(3):157-68. doi: 10.1038/nrmicro2960. Epub 2013 Jan 28. Nat Rev Microbiol. 2013. PMID: 23353768 Free PMC article. Review.
Cited by
- Multidrug-Resistant and Virulent Organisms Trauma Infections: Trauma Infectious Disease Outcomes Study Initiative.
Mende K, Akers KS, Tyner SD, Bennett JW, Simons MP, Blyth DM, Li P, Stewart L, Tribble DR. Mende K, et al. Mil Med. 2022 May 4;187(Suppl 2):42-51. doi: 10.1093/milmed/usab131. Mil Med. 2022. PMID: 35512375 Free PMC article. Review. - Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics.
Lebeaux D, Ghigo JM, Beloin C. Lebeaux D, et al. Microbiol Mol Biol Rev. 2014 Sep;78(3):510-43. doi: 10.1128/MMBR.00013-14. Microbiol Mol Biol Rev. 2014. PMID: 25184564 Free PMC article. Review. - Molecular mechanisms involved in Bacillus subtilis biofilm formation.
Mielich-Süss B, Lopez D. Mielich-Süss B, et al. Environ Microbiol. 2015 Mar;17(3):555-65. doi: 10.1111/1462-2920.12527. Epub 2014 Jul 7. Environ Microbiol. 2015. PMID: 24909922 Free PMC article. Review. - Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms.
Cairns LS, Hobley L, Stanley-Wall NR. Cairns LS, et al. Mol Microbiol. 2014 Aug;93(4):587-98. doi: 10.1111/mmi.12697. Epub 2014 Jul 18. Mol Microbiol. 2014. PMID: 24988880 Free PMC article. Review. - Chance and Necessity in Bacillus subtilis Development.
Mirouze N, Dubnau D. Mirouze N, et al. Microbiol Spectr. 2013 Oct;1(1):10.1128/microbiolspectrum.TBS-0004-2012. doi: 10.1128/microbiolspectrum.TBS-0004-2012. Microbiol Spectr. 2013. PMID: 26184812 Free PMC article.
References
- Berne BJP, R, editors. Dynamic Light Scattering. New York: Wiley; 1976.
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006a;59:1229–1238. - PubMed
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006b;59:1229–1238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM082137/GM/NIGMS NIH HHS/United States
- R01 GM018568/GM/NIGMS NIH HHS/United States
- R01 GM086258/GM/NIGMS NIH HHS/United States
- U54 AI057159/AI/NIAID NIH HHS/United States
- GM82137/GM/NIGMS NIH HHS/United States
- GM58213/GM/NIGMS NIH HHS/United States
- AI057159/AI/NIAID NIH HHS/United States
- GM18568/GM/NIGMS NIH HHS/United States
- R37 GM018568/GM/NIGMS NIH HHS/United States
- R01 GM058213/GM/NIGMS NIH HHS/United States
- GM086258/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases