Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease - PubMed (original) (raw)
. 2012 Jul 15;21(14):3255-63.
doi: 10.1093/hmg/dds165. Epub 2012 Apr 27.
Affiliations
- PMID: 22543974
- PMCID: PMC3384386
- DOI: 10.1093/hmg/dds165
Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease
Scott Smemo et al. Hum Mol Genet. 2012.
Abstract
Recent studies have identified the genetic underpinnings of a growing number of diseases through targeted exome sequencing. However, this strategy ignores the large component of the genome that does not code for proteins, but is nonetheless biologically functional. To address the possible involvement of regulatory variation in congenital heart diseases (CHDs), we searched for regulatory mutations impacting the activity of TBX5, a dosage-dependent transcription factor with well-defined roles in the heart and limb development that has been associated with the Holt-Oram syndrome (heart-hand syndrome), a condition that affects 1/100 000 newborns. Using a combination of genomics, bioinformatics and mouse genetic engineering, we scanned ∼700 kb of the TBX5 locus in search of cis-regulatory elements. We uncovered three enhancers that collectively recapitulate the endogenous expression pattern of TBX5 in the developing heart. We re-sequenced these enhancer elements in a cohort of non-syndromic patients with isolated atrial and/or ventricular septal defects, the predominant cardiac defects of the Holt-Oram syndrome, and identified a patient with a homozygous mutation in an enhancer ∼90 kb downstream of TBX5. Notably, we demonstrate that this single-base-pair mutation abrogates the ability of the enhancer to drive expression within the heart in vivo using both mouse and zebrafish transgenic models. Given the population-wide frequency of this variant, we estimate that 1/100 000 individuals would be homozygous for this variant, highlighting that a significant number of CHD associated with TBX5 dysfunction might arise from non-coding mutations in TBX5 heart enhancers, effectively decoupling the heart and hand phenotypes of the Holt-Oram syndrome.
Figures
Figure 1.
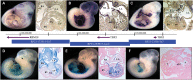
Regulatory landscape of the TBX5 locus. Whole mount and histological sections through the heart of embryonic day 11.5 (E11.5) embryos. Top row: In situ hybridizations showing endogenous expression (purple) of (A) RBM19, (B) TBX5 and (C) TBX3. Bottom row: β-Galactosidase staining (blue) captures the regulatory landscape of enhancers within BACs (D) RP23-173F14-Tn7, (E) RP23-267B15-LacZ and (F) RP23-235J6-Tn7. The genes probed for in A, B and C are contained, respectively, within the BACs tested in D, E and F. There is a 46 bp gap between the BACs containing RBM19 and TBX5, encompassing nucleotides 120 424 690–120 424 646. There is 1140 bp of overlap between the BACs containing TBX5 and TBX3 encompassing nucleotides 120 216 612–120 217 754. Panels B, D–F, forelimb is outlined. A–E, transverse sections; F, sagittal section. A, atrium; RA, right atrium; LA, left atrium; AS, atrial septum; V, ventricle; RV, right ventricle; LV, left ventricle; VS, ventricular septum; AVC, atrioventricular canal.
Figure 2.
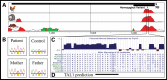
Location of and β-galactosidase expression driven by TBX5 enhancers. (A and B) The TBX5 locus. (C) Individual elements tested for enhancer activity. Sequences with enhancer activity consistent with the expression pattern of TBX5 are dark blue. (D) Pairwise alignments of the mouse genome to human and chicken identify evolutionarily conserved regions (ECRs), which are displayed as peaks. (E–G) Whole mount, isolated hearts and transverse histological sections of transgenic animals carrying individual TBX5 enhancers at E11.5. Consistent expression, across multiple lines, was observed for all three enhancers in the ventricular myocardium, and although it is evident here for ECRs 2 and 16, atrial expression was not recapitulated in independent lines. Only enhancer 2 reproducibly drove expression outside the heart, in the eye. See Fig. 1 for abbreviations and
Supplementary Material, Figs S1–S6
for additional lines and time points.
Figure 3.
Enhancer 9 genomic context and nucleotide conservation. (A) Human alignment to chicken, opossum and mouse at the enhancer 9 region (black bars, top) shows that this enhancer contains two short segments of highly conserved nucleotides (red peaks). (B) One patient possessed a homozygous G→T sequence alteration (hg19 chr12:114 704 515), with two heterozygous parents. (C) The variant nucleotide (boxed) is highly conserved and overlaps a predicted, conserved binding site for TAL1 (D).
Figure 4.
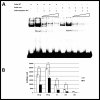
Reduced binding to mutant DNA. Radioactive DNA probe containing wild-type G allele (WT) is bound to a greater extent than probe containing mutant T allele (mut) (A, lanes 1, 2 versus 6, 7; B, 20 and 30 μg). Cold WT probe competitively displaces mutant probe faster than WT (A, lanes 8–10 versus 3–5; B, ×5, ×10 and ×20). Statistical analyses were performed with two-way ANOVA. Each bar represents median ± standard error (_n_= 3). *P< 0.05 versus wild-type.
Figure 5.
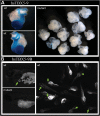
A regulatory variant abrogates enhancer function. (A) A single-base-pair mutation in enhancer 9 abrogates its ability to drive cardiac expression. (B) Enhancer 9B, containing a 515 bp ECR within enhancer 9, drives cardiac expression in zebrafish. When mutated similarly, it too loses enhancer activity.
Similar articles
- KLF13 is a genetic modifier of the Holt-Oram syndrome gene TBX5.
Darwich R, Li W, Yamak A, Komati H, Andelfinger G, Sun K, Nemer M. Darwich R, et al. Hum Mol Genet. 2017 Mar 1;26(5):942-954. doi: 10.1093/hmg/ddx009. Hum Mol Genet. 2017. PMID: 28164238 - Patient-Specific TBX5-G125R Variant Induces Profound Transcriptional Deregulation and Atrial Dysfunction.
van Ouwerkerk AF, Bosada FM, van Duijvenboden K, Houweling AC, Scholman KT, Wakker V, Allaart CP, Uhm JS, Mathijssen IB, Baartscheer T, Postma AV, Barnett P, Verkerk AO, Boukens BJ, Christoffels VM. van Ouwerkerk AF, et al. Circulation. 2022 Feb 22;145(8):606-619. doi: 10.1161/CIRCULATIONAHA.121.054347. Epub 2022 Feb 3. Circulation. 2022. PMID: 35113653 Free PMC article. - TBX5 loss-of-function mutation contributes to atrial fibrillation and atypical Holt-Oram syndrome.
Guo DF, Li RG, Yuan F, Shi HY, Hou XM, Qu XK, Xu YJ, Zhang M, Liu X, Jiang JQ, Yang YQ, Qiu XB. Guo DF, et al. Mol Med Rep. 2016 May;13(5):4349-56. doi: 10.3892/mmr.2016.5043. Epub 2016 Mar 24. Mol Med Rep. 2016. PMID: 27035640 Clinical Trial. - TBX5: A Key Regulator of Heart Development.
Steimle JD, Moskowitz IP. Steimle JD, et al. Curr Top Dev Biol. 2017;122:195-221. doi: 10.1016/bs.ctdb.2016.08.008. Epub 2016 Sep 28. Curr Top Dev Biol. 2017. PMID: 28057264 Free PMC article. Review.
Cited by
- Regulatory de novo mutations underlying intellectual disability.
De Vas MG, Boulet F, Joshi SS, Garstang MG, Khan TN, Atla G, Parry D, Moore D, Cebola I, Zhang S, Cui W, Lampe AK, Lam WW; Genomics England Research Consortium; Ferrer J, Pradeepa MM, Atanur SS. De Vas MG, et al. Life Sci Alliance. 2023 Feb 28;6(5):e202201843. doi: 10.26508/lsa.202201843. Print 2023 May. Life Sci Alliance. 2023. PMID: 36854624 Free PMC article. - The clinical and genetic characteristics of permanent neonatal diabetes (PNDM) in the state of Qatar.
Al-Khawaga S, Mohammed I, Saraswathi S, Haris B, Hasnah R, Saeed A, Almabrazi H, Syed N, Jithesh P, El Awwa A, Khalifa A, AlKhalaf F, Petrovski G, Abdelalim EM, Hussain K. Al-Khawaga S, et al. Mol Genet Genomic Med. 2019 Oct;7(10):e00753. doi: 10.1002/mgg3.753. Epub 2019 Aug 23. Mol Genet Genomic Med. 2019. PMID: 31441606 Free PMC article. - PTPN11 Gene Mutations and Its Association with the Risk of Congenital Heart Disease.
Xu ZQ, Chen WC, Li YJ, Suo MJ, Tian GX, Sheng W, Huang GY. Xu ZQ, et al. Dis Markers. 2022 Apr 9;2022:8290779. doi: 10.1155/2022/8290779. eCollection 2022. Dis Markers. 2022. PMID: 35440950 Free PMC article. Retracted. - Regulatory variation within 3'UTR of STAT5A correlates with sudden cardiac death in Chinese populations.
Yu H, Guo Y, Yang Z, Zhang Q, Xu J, Yang Q, Qu Y, Tan R, Li L, He Y, Li C, Zhang S, Luo B, Gao Y. Yu H, et al. Forensic Sci Res. 2021 Jul 23;7(4):726-735. doi: 10.1080/20961790.2021.1895410. eCollection 2022. Forensic Sci Res. 2021. PMID: 37101540 Free PMC article. - The role of histone modification and a regulatory single-nucleotide polymorphism (rs2071166) in the Cx43 promoter in patients with TOF.
Gu R, Xu J, Lin Y, Sheng W, Ma D, Ma X, Huang G. Gu R, et al. Sci Rep. 2017 Sep 5;7(1):10435. doi: 10.1038/s41598-017-10756-6. Sci Rep. 2017. PMID: 28874875 Free PMC article.
References
- Hoffman J.I., Kaplan S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 2002;39:1890–1900. - PubMed
- Basson C.T., Bachinsky D.R., Lin R.C., Levi T., Elkins J.A., Soults J., Grayzel D., Kroumpouzou E., Traill T.A., Leblanc-Straceski J., et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt–Oram syndrome. Nat. Genet. 1997;15:30–35. - PubMed
- Garg V., Kathiriya I.S., Barnes R., Schluterman M.K., King I.N., Butler C.A., Rothrock C.R., Eapen R.S., Hirayama-Yamada K., Joo K., et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. - PubMed
- Schott J.J., Benson D.W., Basson C.T., Pease W., Silberbach G.M., Moak J.P., Maron B.J., Seidman C.E., Seidman J.G. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. - PubMed
- Lopes Floro K., Artap S.T., Preis J.I., Fatkin D., Chapman G., Furtado M.B., Harvey R.P., Hamada H., Sparrow D.B., Dunwoodie S.L. Loss of Cited2 causes congenital heart disease by perturbing left-right patterning of the body axis. Hum. Mol. Genet. 2011;20:1097–1110. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- HL088393/HL/NHLBI NIH HHS/United States
- R01 HL092153/HL/NHLBI NIH HHS/United States
- HG004428/HG/NHGRI NIH HHS/United States
- R01 HL088393/HL/NHLBI NIH HHS/United States
- DK078871/DK/NIDDK NIH HHS/United States
- T32GM007197/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases