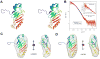Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata - PubMed (original) (raw)
doi: 10.1002/prot.24102. Epub 2012 Jun 4.
Seung Joong Kim, Danalyn Manglicmot, Kevin T Bain, Jeremiah Gilmore, Tarun Gheyi, Jeremy Phillips, Ursula Pieper, Javier Fernandez-Martinez, Josef D Franke, Tsutomu Matsui, Hiro Tsuruta, Shane Atwell, Devon A Thompson, J Spencer Emtage, Stephen R Wasserman, Michael P Rout, Andrej Sali, J Michael Sauder, Steven C Almo, Stephen K Burley
Affiliations
- PMID: 22544723
- PMCID: PMC3686472
- DOI: 10.1002/prot.24102
Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata
Parthasarathy Sampathkumar et al. Proteins. 2012 Aug.
Abstract
The nuclear pore complex (NPC), embedded in the nuclear envelope, is a large, dynamic molecular assembly that facilitates exchange of macromolecules between the nucleus and the cytoplasm. The yeast NPC is an eightfold symmetric annular structure composed of ~456 polypeptide chains contributed by ~30 distinct proteins termed nucleoporins. Nup116, identified only in fungi, plays a central role in both protein import and mRNA export through the NPC. Nup116 is a modular protein with N-terminal "FG" repeats containing a Gle2p-binding sequence motif and a NPC targeting domain at its C-terminus. We report the crystal structure of the NPC targeting domain of Candida glabrata Nup116, consisting of residues 882-1034 [CgNup116(882-1034)], at 1.94 Å resolution. The X-ray structure of CgNup116(882-1034) is consistent with the molecular envelope determined in solution by small-angle X-ray scattering. Structural similarities of CgNup116(882-1034) with homologous domains from Saccharomyces cerevisiae Nup116, S. cerevisiae Nup145N, and human Nup98 are discussed.
Copyright © 2012 Wiley Periodicals, Inc.
Figures
Figure 1
A: Stereoview of the CgNup116(882-1034) monomer. Cartoon of Chain A is shown as a rainbow from blue to red from N- to C-terminus. B: Comparison of the merged experimental SAXS profile (red) of CgNup116(882-1034) with the SAXS profile computed by IMP FoXS (blue) from the complete monomer model of CgNup116(882-1034). Inset shows the SAXS profiles in the Guinier plot, with a Rg fit of 18.38±0.24 Å. Dmax of radial distribution function, P(r), is 60.65 Å. C and D: Comparison of the shapes of CgNup116(882-1034) (represented as a mesh) calculated from the experimental SAXS profile by GASBOR (C) and SASTBX (D).
Figure 2
A: Structure based sequence alignment of the structures of CgNup116(882-1034, PDB Code 3NF5), ScNup116 bound to ScNup82:ScNup159 complex (PDB Code 3PBP), autoproteolytic domain of ScNup145 (PDB Code 3KEP), and autoproteolytic domain of HsNup98 (PDB Code 2Q5X). Secondary structural elements of CgNup116(882-1034) and ScNup116 bound to ScNup82:ScNup159 complex are displayed in green and black, respectively. Residues of ScNup116 contributing to its binary interaction with ScNup82 β-propeller domain are marked with blue star. Sites of autoproteolysis of ScNup145N and HsNup98 are indicated with an orange circle. The Ser residue at the autoproteolytic site of ScNup145N and HsNup98 are replaced by Glu and Ala in proteins used for structure determination, respectively. B: Stereoview of CgNup116(882-1034; green) superposed on the structure of ScNup116 bound to ScNup82:ScNup159 complex (grey). Helix α4 of CgNup116 is structurally equivalent to the helix αB of ScNup116. C: Stereoview of the CgNup116(882-1034; green) superposed on the autoproteolytic domain of ScNup145 (grey). D: Stereoview of the CgNup116(882-1034; green) superposed on the autoproteolytic domain of HsNup98 (grey). Residues preceding the β1-strands are highlighted in magenta and orange for CgNup116(882-1034) and HsNup98, respectively. The N- and C-terminal residues are shown on the cartoons as blue and red spheres, respectively.
Similar articles
- Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex.
Yoshida K, Seo HS, Debler EW, Blobel G, Hoelz A. Yoshida K, et al. Proc Natl Acad Sci U S A. 2011 Oct 4;108(40):16571-6. doi: 10.1073/pnas.1112846108. Epub 2011 Sep 19. Proc Natl Acad Sci U S A. 2011. PMID: 21930948 Free PMC article. - Architecture of the linker-scaffold in the nuclear pore.
Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A. Petrovic S, et al. Science. 2022 Jun 10;376(6598):eabm9798. doi: 10.1126/science.abm9798. Epub 2022 Jun 10. Science. 2022. PMID: 35679425 Free PMC article. - The integral membrane protein snl1p is genetically linked to yeast nuclear pore complex function.
Ho AK, Raczniak GA, Ives EB, Wente SR. Ho AK, et al. Mol Biol Cell. 1998 Feb;9(2):355-73. doi: 10.1091/mbc.9.2.355. Mol Biol Cell. 1998. PMID: 9450961 Free PMC article. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review. - Structural analysis of the nuclear pore complex by integrated approaches.
Elad N, Maimon T, Frenkiel-Krispin D, Lim RY, Medalia O. Elad N, et al. Curr Opin Struct Biol. 2009 Apr;19(2):226-32. doi: 10.1016/j.sbi.2009.02.009. Epub 2009 Mar 25. Curr Opin Struct Biol. 2009. PMID: 19327984 Review.
Cited by
- Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform.
Fernandez-Martinez J, Kim SJ, Shi Y, Upla P, Pellarin R, Gagnon M, Chemmama IE, Wang J, Nudelman I, Zhang W, Williams R, Rice WJ, Stokes DL, Zenklusen D, Chait BT, Sali A, Rout MP. Fernandez-Martinez J, et al. Cell. 2016 Nov 17;167(5):1215-1228.e25. doi: 10.1016/j.cell.2016.10.028. Epub 2016 Nov 10. Cell. 2016. PMID: 27839866 Free PMC article. - Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Upla P, Kim SJ, Sampathkumar P, Dutta K, Cahill SM, Chemmama IE, Williams R, Bonanno JB, Rice WJ, Stokes DL, Cowburn D, Almo SC, Sali A, Rout MP, Fernandez-Martinez J. Upla P, et al. Structure. 2017 Mar 7;25(3):434-445. doi: 10.1016/j.str.2017.01.006. Epub 2017 Feb 2. Structure. 2017. PMID: 28162953 Free PMC article. - Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions.
Raices M, D'Angelo MA. Raices M, et al. Nat Rev Mol Cell Biol. 2012 Nov;13(11):687-99. doi: 10.1038/nrm3461. Nat Rev Mol Cell Biol. 2012. PMID: 23090414 Review. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review. - Integrative structure and functional anatomy of a nuclear pore complex.
Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, Chemmama IE, Pellarin R, Echeverria I, Shivaraju M, Chaudhury AS, Wang J, Williams R, Unruh JR, Greenberg CH, Jacobs EY, Yu Z, de la Cruz MJ, Mironska R, Stokes DL, Aitchison JD, Jarrold MF, Gerton JL, Ludtke SJ, Akey CW, Chait BT, Sali A, Rout MP. Kim SJ, et al. Nature. 2018 Mar 22;555(7697):475-482. doi: 10.1038/nature26003. Epub 2018 Mar 14. Nature. 2018. PMID: 29539637 Free PMC article.
References
- Cronshaw JM, Matunis JM. The nuclear pore complex: disease associations and functional correlations. Trends in Endocrinology and Metabolism. 2004;15:34–39. - PubMed
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang WH, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM083960/GM/NIGMS NIH HHS/United States
- P41RR001209/RR/NCRR NIH HHS/United States
- U54 GM074945/GM/NIGMS NIH HHS/United States
- U54 GM103511/GM/NIGMS NIH HHS/United States
- U54 GM094662/GM/NIGMS NIH HHS/United States
- R01 GM062427/GM/NIGMS NIH HHS/United States
- U01 GM098256/GM/NIGMS NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- P41 RR001209/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases