Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice - PubMed (original) (raw)
Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice
Andres Gomez et al. PLoS One. 2012.
Abstract
Background: HLA-DRB1 0401 is associated with susceptibility, while HLA-DRB1 0402 is associated with resistance to developing rheumatoid arthritis (RA) and collagen-induced arthritis in humans and transgenic mice respectively. The influence of gut-joint axis has been suggested in RA, though not yet proven.
Methodology/principal findings: We have used HLA transgenic mice carrying arthritis susceptible and -resistant HLA-DR genes to explore if genetic factors and their interaction with gut flora gut can be used to predict susceptibility to develop arthritis. Pyrosequencing of the 16S rRNA gene from the fecal microbiomes of DRB1 0401 and DRB1 0402 transgenic mice revealed that the guts of 0401 mice is dominated by a Clostridium-like bacterium, whereas the guts of 0402 mice are enriched for members of the Porphyromonadaceae family and Bifidobacteria. DRB1 0402 mice harbor a dynamic sex and age-influenced gut microbiome while DRB1 0401 mice did not show age and sex differences in gut microbiome even though they had altered gut permeability. Cytokine transcripts, measured by rtPCR, in jejuna showed differential TH17 regulatory network gene transcripts in 0401 and 0402 mice.
Conclusions/significance: We have demonstrated for the first time that HLA genes in association with the gut microbiome may determine the immune environment and that the gut microbiome might be a potential biomarker as well as contributor for susceptibility to arthritis. Identification of pathogenic commensal bacteria would provide new understanding of disease pathogenesis, thereby leading to novel approaches for therapy.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
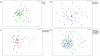
Figure 1. Multidimensional Scalinge analysis of the fecal microbiomes of arthritis-susceptible *0401 and –resistant *0402 mice.
16S-rDNA bacterial community structure differences can be visualized with each symbol representing data from a single mouse fecal sample. (a) 16S-rDNA bacterial community structures between *0401 (n = 41) and *0402 (n = 45) mice do not differ significantly (ANOSIM R = 0.14). (b) *0401 (n = 22) and *0402 (n = 21) males do not show significant differences in fecal microbiome structure (ANOSIM R = 0.14), while (c) fecal microbiomes of *0401 (n = 19) and *0402 (n = 24) females differ significantly (ANOSIM R = 0.436). (d) Shaded area shows that the fecal microbiomes of *0402 females are compact and may be driving differences between both genotypes.
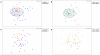
Figure 2. Sex and Age based Multidimensional Scalinge analysis of fecal microbiomes.
(a) *0402 mice show significantly different fecal microbiome structure according to sex (ANOSIM R = 0.302) and (b) age (ANOSIM R = 0.423). (c) *0401 mice lost sex and (d) age-based differences in fecal microbiome (ANOSIM R = 0.052 and R = 0.043 respectively). Shaded areas show compact microbiome structures.
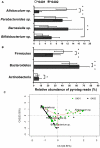
Figure 3. Relative abundance of OTUs in the fecal microbiomes of *0401 and *0402 mice.
(a) Allobaculum sp. is is the most abundant OTU in the *0401 mice, while Barnesiella sp. occurs with highest frequency in *0402 mice (P<0.05). (b) Similar taxa distributions are observed at phyla level with Bacteroidetes: Firmicutes ratios more even in *0401 (n = 41) compared to *0402 (n = 45) mice. Data are presented as mean ± S.E., *P<0.05, **P<0.01, ***P<0.001. (c) Correspondence analysis plot shows the degree of correlation between specific OTUs and mice genotype. The red vectors point to the center of gravity of the samples where each OTU mostly occurs. The distance between the tip of the vector and the samples (dots) give an indication of the probability of OTU content in each sample. Green and black dots represent *0401 and *0402 fecal samples respectively. Percentages in parentheses in the CA plots describe the amount of variation explained by each axis.
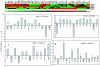
Figure 4. Sex based relative abundance of OTUs in the fecal microbiomes of *0401 and *0402 mice.
(a) *0402 females (n = 24) show significantly higher relative abundances of _Bifidobacterium_-Parabacteroides OTUs compared to males (n = 21), whose microbiomes present significantly higher levels of Barnesiella viscericola. (b) Correspondence analysis plot displays sex-based correlation between OTUs in *0402 mice. (c) Significantly higher relative abundances of Bifidobacterium sp. were observed in *0401 males (n = 22) compared to females (n = 19), (d) despite loss of dynamic sex based differences in the fecal microbiomes of *0401 mice. Percentages in parentheses in the CA plots describe the amount of variation explained by each axis. Data are presented as mean ± S.E. *P<0.05, **P<0.01, ***P<0.001.
Figure 5. Gut permeability.
(a) Transgenic arthritic mice showed significantly higher gut permeability compared to naïve mice (n = 5 each group). (b) Intestinal permeability in naïve *0401 and *0402 transgenic male and female mice at >4 and <4 months of age. *0401F<4 mo vs >4 mo, P<0.04; *0401F vs *0402F >4 mo, P<0.03 (n = 5–8 in groups).
Figure 6. A)) Heat map showing expression levels of cytokines and chemokine transcripts in jejenum of *0401 and *0402 male and female mice (n = 3 in each group).
(b) Comparison of fold change in gene transcript levels between *0401 females and males, (c) *0402 females and males, (d) females of each genotype and (e) males of each genotype. Results are given as fold-changes of mean copy-numbers relative to the mean copy-numbers of the comparative group. *P<0.05 and **P<0.01 and more. Data points with 3 or more fold differences and significance of more than P<0.05 are shown.
Figure 7. Role of the gut microbiome in susceptibility to arthritis.
HLA genotype may shape the gut microbiome in an individual. The DRB1*0401 gene associated with predisposition to Rheumatoid arthritis, may induce a lower gut Bacteroidetes: Firmicutes ratio compared to that shaped by the DRB1*0402 gene, known to be associated with resistance to arthritis. This model suggests that a dysbiotic or arthritogenic gut microbiome may be dominated by a Clostridia-like bacterium (Firmicutes phylum) in susceptible individuals, while competent/tolerant immune responses are enhanced by increased abundances of Bifidobacterium spp. in resistance to RA. The gut microbiome has a crucial influence on maintaining homeostasis of the gut immune system by predicting pro-inflammatory (TH1/Th17) or anti-inflammatory (TH1/TH2) responses. Environmental triggers like smoking, diet and infectious agents along with sex hormones and age-dependent changes, may further modulate the gut immune system and enhance pro-inflammatory conditions in genetically susceptible individuals. In synthesis, an overall/systemic immune response generated by innate immune cells, may be originated at gut level and this response may be regulated by the gut microbiome via HLA genotype. This chain of events may determine the onset of autoimmune diseases like rheumatoid arthritis.
Similar articles
- Bugs & us: the role of the gut in autoimmunity.
Luckey D, Gomez A, Murray J, White B, Taneja V. Luckey D, et al. Indian J Med Res. 2013 Nov;138(5):732-43. Indian J Med Res. 2013. PMID: 24434325 Free PMC article. Review. - An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis.
Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, Nelson H, Matteson EL, Taneja V. Chen J, et al. Genome Med. 2016 Apr 21;8(1):43. doi: 10.1186/s13073-016-0299-7. Genome Med. 2016. PMID: 27102666 Free PMC article. - Suppression of Inflammatory Arthritis by Human Gut-Derived Prevotella histicola in Humanized Mice.
Marietta EV, Murray JA, Luckey DH, Jeraldo PR, Lamba A, Patel R, Luthra HS, Mangalam A, Taneja V. Marietta EV, et al. Arthritis Rheumatol. 2016 Dec;68(12):2878-2888. doi: 10.1002/art.39785. Arthritis Rheumatol. 2016. PMID: 27337150 Free PMC article. - Cytokines pre-determined by genetic factors are involved in pathogenesis of Rheumatoid arthritis.
Taneja V. Taneja V. Cytokine. 2015 Oct;75(2):216-21. doi: 10.1016/j.cyto.2014.11.028. Epub 2014 Dec 23. Cytokine. 2015. PMID: 25541434 Free PMC article. - To B or not to B: role of B cells in pathogenesis of arthritis in HLA transgenic mice.
Behrens M, Smart M, Luckey D, Luthra H, Taneja V. Behrens M, et al. J Autoimmun. 2011 Sep;37(2):95-103. doi: 10.1016/j.jaut.2011.05.002. Epub 2011 Jun 12. J Autoimmun. 2011. PMID: 21665435 Free PMC article. Review.
Cited by
- Immunogenetic control of the intestinal microbiota.
Marietta E, Rishi A, Taneja V. Marietta E, et al. Immunology. 2015 Jul;145(3):313-22. doi: 10.1111/imm.12474. Epub 2015 Jun 3. Immunology. 2015. PMID: 25913295 Free PMC article. Review. - Gut microbiota and rheumatoid arthritis: From pathogenesis to novel therapeutic opportunities.
Zhao T, Wei Y, Zhu Y, Xie Z, Hai Q, Li Z, Qin D. Zhao T, et al. Front Immunol. 2022 Sep 8;13:1007165. doi: 10.3389/fimmu.2022.1007165. eCollection 2022. Front Immunol. 2022. PMID: 36159786 Free PMC article. Review. - Microbiota-assisted therapy for systemic inflammatory arthritis: advances and mechanistic insights.
Li B, Yang B, Liu X, Zhao J, Ross RP, Stanton C, Zhang H, Chen W. Li B, et al. Cell Mol Life Sci. 2022 Aug 6;79(9):470. doi: 10.1007/s00018-022-04498-6. Cell Mol Life Sci. 2022. PMID: 35932328 Free PMC article. Review. - A gut feeling about arthritis.
Mathis D. Mathis D. Elife. 2013 Nov 5;2:e01608. doi: 10.7554/eLife.01608. Elife. 2013. PMID: 24192040 Free PMC article. - The Emerging World of Microbiome in Autoimmune Disorders: Opportunities and Challenges.
Mangalam AK, Yadav M, Yadav R. Mangalam AK, et al. Indian J Rheumatol. 2021 Mar;16(1):57-72. doi: 10.4103/injr.injr_210_20. Epub 2021 Mar 23. Indian J Rheumatol. 2021. PMID: 34531642 Free PMC article.
References
- Klareskog L, Padyukov L, Lorentzen J, Alfredsson L. Mechanisms of disease: Genetic susceptibility and environmental triggers in the development of rheumatoid arthritis. Nat Clin Pract Rheumatol. 2006;2:425–433. - PubMed
- Moen K, Brun JG, Valen M, Skartveit L, Eribe EK, et al. Synovial inflammation in active rheumatoid arthritis and psoriatic arthritis facilitates trapping of a variety of oral bacterial DNAs. Clin Exp Rheumatol. 2006;24:656–663. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials