Disruption of protein quality control in Parkinson's disease - PubMed (original) (raw)
Review
Disruption of protein quality control in Parkinson's disease
Casey Cook et al. Cold Spring Harb Perspect Med. 2012 May.
Abstract
Parkinson's disease (PD), like a number of neurodegenerative diseases associated with aging, is characterized by the abnormal accumulation of protein in a specific subset of neurons. Although researchers have recently elucidated the genetic causes of PD, much remains unknown about what causes increased protein deposition in the disease. Given that increased protein aggregation may result not only from an increase in production, but also from decreased protein clearance, it is imperative to investigate both possibilities as potential PD culprits. This article provides a review of the systems that regulate protein clearance, including the ubiquitin proteasome system (UPS) and the autophagy-lysosomal pathway. Literature implicating failure of these mechanisms-such as UPS dysfunction resulting from environmental toxins and mutations in α-synuclein and parkin, as well as macroautophagic pathway failure because of oxidative stress and aging-in the pathogenesis of PD is also discussed.
Figures
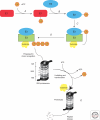
Figure 1.
Representation of ubiquitin-proteasome proteolysis. Ubiquitin is first activated by the ubiquitin-activating enzyme E1 (A), an ubiquitin-carrier protein, E2, and ATP. The product of this reaction is a high-energy E2 ∼ ubiquitin thiol ester intermediate (B). Protein substrates are then ubiquitinated by either binding of the substrate to a specific ubiquitin-protein ligase (E3), and then the E2-bound activated ubiquitin is transferred directly to the E3-bound protein substrate. Or alternatively, the activated ubiquitin can be transferred from the E2 to the E3, prior to its conjugation to the E3-bound substrate (C). Following conjugation of the first ubiquitin molecule to the protein substrate, additional ubiquitin molecules can be added to the internal lysine residues of ubiquitin to form a polyubiquitin chain on the substrate (D). The ubiquitinated substrate is then recognized and degraded by the 26S proteasome complex, leading to the release of short peptides (E). Ubiquitin is recycled via the activity of deubiquitinating enzymes.
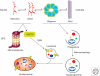
Figure 2.
Impact of aggregation on mechanisms of protein degradation. Soluble misfolded monomers and dimers can be recognized by both the UPS or CMA-related chaperones, and subsequently degraded by either of these two pathways. In the case of CMA, cytosolic proteins (i.e., α-synuclein) are recognized by a chaperone (i.e., Hsc70), which delivers the target protein to the lysosome via a receptor protein present in the lysosomal membrane. However, on more complex assembly (oligomer or fibril formation) of the target protein, macroautophagy is the only mechanism available to clear the more insoluble and highly ordered aggregates.
Similar articles
- Environmental neurotoxic chemicals-induced ubiquitin proteasome system dysfunction in the pathogenesis and progression of Parkinson's disease.
Sun F, Kanthasamy A, Anantharam V, Kanthasamy AG. Sun F, et al. Pharmacol Ther. 2007 Jun;114(3):327-44. doi: 10.1016/j.pharmthera.2007.04.001. Epub 2007 Apr 19. Pharmacol Ther. 2007. PMID: 17521740 Review. - Age-related dysfunctions of the autophagy lysosomal pathway in hippocampal pyramidal neurons under proteasome stress.
Gavilán E, Pintado C, Gavilan MP, Daza P, Sánchez-Aguayo I, Castaño A, Ruano D. Gavilán E, et al. Neurobiol Aging. 2015 May;36(5):1953-63. doi: 10.1016/j.neurobiolaging.2015.02.025. Epub 2015 Feb 28. Neurobiol Aging. 2015. PMID: 25817083 - Protein degradation pathways in Parkinson's disease: curse or blessing.
Ebrahimi-Fakhari D, Wahlster L, McLean PJ. Ebrahimi-Fakhari D, et al. Acta Neuropathol. 2012 Aug;124(2):153-72. doi: 10.1007/s00401-012-1004-6. Epub 2012 Jun 29. Acta Neuropathol. 2012. PMID: 22744791 Free PMC article. Review. - Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies.
Ciechanover A, Kwon YT. Ciechanover A, et al. Exp Mol Med. 2015 Mar 13;47(3):e147. doi: 10.1038/emm.2014.117. Exp Mol Med. 2015. PMID: 25766616 Free PMC article. Review.
Cited by
- Saposin C, Key Regulator in the Alpha-Synuclein Degradation Mediated by Lysosome.
Ruz C, Barrero FJ, Pelegrina J, Bandrés-Ciga S, Vives F, Duran R. Ruz C, et al. Int J Mol Sci. 2022 Oct 9;23(19):12004. doi: 10.3390/ijms231912004. Int J Mol Sci. 2022. PMID: 36233303 Free PMC article. - A novel LINC00943/miR-671-5p/ELAVL1 ceRNA crosstalk regulates MPP+ toxicity in SK-N-SH cells.
Zhang X, Luan N, Shi J. Zhang X, et al. Metab Brain Dis. 2022 Oct;37(7):2349-2362. doi: 10.1007/s11011-022-01034-0. Epub 2022 Jul 2. Metab Brain Dis. 2022. PMID: 35779150 - Autophagy Stimulation Decreases Dopaminergic Neuronal Death Mediated by Oxidative Stress.
Ramirez-Moreno MJ, Duarte-Jurado AP, Gopar-Cuevas Y, Gonzalez-Alcocer A, Loera-Arias MJ, Saucedo-Cardenas O, Montes de Oca-Luna R, Rodriguez-Rocha H, Garcia-Garcia A. Ramirez-Moreno MJ, et al. Mol Neurobiol. 2019 Dec;56(12):8136-8156. doi: 10.1007/s12035-019-01654-1. Epub 2019 Jun 13. Mol Neurobiol. 2019. PMID: 31197654 - Decreased proteasomal activity causes photoreceptor degeneration in mice.
Ando R, Noda K, Tomaru U, Kamoshita M, Ozawa Y, Notomi S, Hisatomi T, Noda M, Kanda A, Ishibashi T, Kasahara M, Ishida S. Ando R, et al. Invest Ophthalmol Vis Sci. 2014 Jul 3;55(7):4682-90. doi: 10.1167/iovs.13-13272. Invest Ophthalmol Vis Sci. 2014. PMID: 24994871 Free PMC article. - Overexpression of alpha-synuclein at non-toxic levels increases dopaminergic cell death induced by copper exposure via modulation of protein degradation pathways.
Anandhan A, Rodriguez-Rocha H, Bohovych I, Griggs AM, Zavala-Flores L, Reyes-Reyes EM, Seravalli J, Stanciu LA, Lee J, Rochet JC, Khalimonchuk O, Franco R. Anandhan A, et al. Neurobiol Dis. 2015 Sep;81:76-92. doi: 10.1016/j.nbd.2014.11.018. Epub 2014 Dec 8. Neurobiol Dis. 2015. PMID: 25497688 Free PMC article.
References
- Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper JM, Caballero C, Ferrer I, Obeso JA, Schapira AH 2010. Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol 67: 1464–1472 - PubMed
- Anderson JP, Walker DE, Goldstein JM, de Laat R, Banducci K, Caccavello RJ, Barbour R, Huang J, Kling K, Lee M, et al. 2006. Phosphorylation of Ser-129 is the dominant pathological modification of α-synuclein in familial and sporadic Lewy body disease. J Biol Chem 281: 29739–29752 - PubMed
- Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM 2002. Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science 295: 865–868 - PubMed
- Balch WE, Morimoto RI, Dillin A, Kelly JW 2008. Adapting proteostasis for disease intervention. Science 319: 916–919 - PubMed
- Bandyopadhyay U, Cuervo AM 2007. Chaperone-mediated autophagy in aging and neurodegeneration: Lessons from α-synuclein. Exp Gerontol 42: 120–128 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical