Melanesian blond hair is caused by an amino acid change in TYRP1 - PubMed (original) (raw)
Melanesian blond hair is caused by an amino acid change in TYRP1
Eimear E Kenny et al. Science. 2012.
Abstract
Naturally blond hair is rare in humans and found almost exclusively in Europe and Oceania. Here, we identify an arginine-to-cysteine change at a highly conserved residue in tyrosinase-related protein 1 (TYRP1) as a major determinant of blond hair in Solomon Islanders. This missense mutation is predicted to affect catalytic activity of TYRP1 and causes blond hair through a recessive mode of inheritance. The mutation is at a frequency of 26% in the Solomon Islands, is absent outside of Oceania, represents a strong common genetic effect on a complex human phenotype, and highlights the importance of examining genetic associations worldwide.
Figures
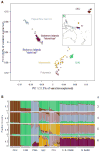
Figure 1
PCA (A) and ADMIXTURE (B) plots demonstrating the genetic relationships between Solomon Islanders (blond and dark hair) genotyped in the present study and several other populations. In PC space, Solomon Islanders are found between New Guineans and Asians, which is consistent with their population history (7). Blond and dark-haired Solomon Islanders show no systematic differences in their ancestry suggesting that blond hair is unlikely to be due to gene flow from other populations (e.g. Europeans). (CEU = Europeans from HapMap; CHB = Han Chinese from HapMap; PNG = Papua New Guinea; MIC = Micronesians; POL = Polynesians; SI = Solomon Islanders).
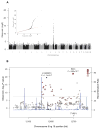
Figure 2
Mapping the blond hair locus in Solomon Islanders. (A) A Manhattan plot and QQ-plot (inset) of the association scores comparing blond and dark haired individuals. The grey line indicates the genome-wide threshold for statistical significance. The QQ-plot shows the distribution of p-values before (black points; λ=1.02) and after (grey points; λ=1.00) removal of the 151 SNPs in the associated 9p23 region. (B) A regional plot of the GWA including the R93C allele, where the signal for the top SNP on the GWA array (blue diamond) and R93C (large red diamond) are indicated. The degree of redness of all SNP’s in the region indicates linkage disequilibrium with R93C. Also shown is recombination rate (navy line) at the signal peak in a window of ± 500 kb around the top GWA SNP. Positions, recombination rates and gene annotations are according to NCBI build 37 (hg 19).
Figure 3
Hair and skin pigmentation variants in TYRP1 (A) Top; a schema of the protein domains of TYRP1 (signal peptide (SI); epidermal growth factor (EGF), copper-binding and transmembrane (TM) domains) indicating variants in humans and other vertebrates that result in lightened skin and/or hair/coat (in brown) or albinism (in grey). Bottom; homology at the first 100 amino acids of TYRP1 in humans and other vertebrates. The location of the mouse brownlight mutation (R38C) and the Solomon Islands blond hair mutation (R93C) are given. (B) Boxplot showing the effect of the R93C genotypes on hair pigmentation in 921 Solomon Islanders.
Similar articles
- The rs387907171 SNP in TYRP1 is not associated with blond hair color on the Island of Bougainville.
Norton HL, Hanna M, Werren E, Friedlaender J. Norton HL, et al. Am J Hum Biol. 2016 May;28(3):431-5. doi: 10.1002/ajhb.22795. Epub 2015 Oct 9. Am J Hum Biol. 2016. PMID: 26450459 - Distribution of an allele associated with blond hair color across Northern Island Melanesia.
Norton HL, Correa EA, Koki G, Friedlaender JS. Norton HL, et al. Am J Phys Anthropol. 2014 Apr;153(4):653-62. doi: 10.1002/ajpa.22466. Epub 2014 Jan 22. Am J Phys Anthropol. 2014. PMID: 24449225 - TYRP1:c.555T>G is a recurrent mutation found in Australian Shepherd and Miniature American Shepherd dogs.
Jancuskova T, Langevin M, Pekova S. Jancuskova T, et al. Anim Genet. 2018 Oct;49(5):500-501. doi: 10.1111/age.12709. Epub 2018 Aug 14. Anim Genet. 2018. PMID: 30109695 No abstract available. - Structure and Function of Human Tyrosinase and Tyrosinase-Related Proteins.
Lai X, Wichers HJ, Soler-Lopez M, Dijkstra BW. Lai X, et al. Chemistry. 2018 Jan 2;24(1):47-55. doi: 10.1002/chem.201704410. Epub 2017 Nov 28. Chemistry. 2018. PMID: 29052256 Review. - Tyrp1 and oculocutaneous albinism type 3.
Sarangarajan R, Boissy RE. Sarangarajan R, et al. Pigment Cell Res. 2001 Dec;14(6):437-44. doi: 10.1034/j.1600-0749.2001.140603.x. Pigment Cell Res. 2001. PMID: 11775055 Review.
Cited by
- Polymorphisms associated with a tropical climate and root crop diet induce susceptibility to metabolic and cardiovascular diseases in Solomon Islands.
Furusawa T, Naka I, Yamauchi T, Natsuhara K, Eddie R, Kimura R, Nakazawa M, Ishida T, Ohtsuka R, Ohashi J. Furusawa T, et al. PLoS One. 2017 Mar 2;12(3):e0172676. doi: 10.1371/journal.pone.0172676. eCollection 2017. PLoS One. 2017. PMID: 28253292 Free PMC article. - Networks Underpinning Symbiosis Revealed Through Cross-Species eQTL Mapping.
Guo Y, Fudali S, Gimeno J, DiGennaro P, Chang S, Williamson VM, Bird DM, Nielsen DM. Guo Y, et al. Genetics. 2017 Aug;206(4):2175-2184. doi: 10.1534/genetics.117.202531. Epub 2017 Jun 22. Genetics. 2017. PMID: 28642272 Free PMC article. - A study in scarlet: MC1R as the main predictor of red hair and exemplar of the flip-flop effect.
Zorina-Lichtenwalter K, Lichtenwalter RN, Zaykin DV, Parisien M, Gravel S, Bortsov A, Diatchenko L. Zorina-Lichtenwalter K, et al. Hum Mol Genet. 2019 Jun 15;28(12):2093-2106. doi: 10.1093/hmg/ddz018. Hum Mol Genet. 2019. PMID: 30657907 Free PMC article. - Global patterns of diversity and selection in human tyrosinase gene.
Hudjashov G, Villems R, Kivisild T. Hudjashov G, et al. PLoS One. 2013 Sep 11;8(9):e74307. doi: 10.1371/journal.pone.0074307. eCollection 2013. PLoS One. 2013. PMID: 24040225 Free PMC article. - Minimal Efficacy of Nitisinone Treatment in a Novel Mouse Model of Oculocutaneous Albinism, Type 3.
Onojafe IF, Megan LH, Melch MG, Aderemi JO, Alur RP, Abu-Asab MS, Chan CC, Bernardini IM, Albert JS, Cogliati T, Adams DR, Brooks BP. Onojafe IF, et al. Invest Ophthalmol Vis Sci. 2018 Oct 1;59(12):4945-4952. doi: 10.1167/iovs.16-20293. Invest Ophthalmol Vis Sci. 2018. PMID: 30347088 Free PMC article.
References
- Sturm RA. Molecular genetics of human pigmentation diversity. Hum Mol Genet. 2009 Apr 15;18:R9. - PubMed
- Norton HL, et al. Skin and hair pigmentation variation in Island Melanesia. Am J Phys Anthropol. 2006 Jun;130:254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 ES015794/ES/NIEHS NIH HHS/United States
- R01 HL078885/HL/NHLBI NIH HHS/United States
- G0600705/MRC_/Medical Research Council/United Kingdom
- U19 AI077439/AI/NIAID NIH HHS/United States
- R01 HL088133/HL/NHLBI NIH HHS/United States
- RD1634/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials