Removal of shelterin reveals the telomere end-protection problem - PubMed (original) (raw)
Removal of shelterin reveals the telomere end-protection problem
Agnel Sfeir et al. Science. 2012.
Abstract
The telomere end-protection problem is defined by the aggregate of DNA damage signaling and repair pathways that require repression at telomeres. To define the end-protection problem, we removed the whole shelterin complex from mouse telomeres through conditional deletion of TRF1 and TRF2 in nonhomologous end-joining (NHEJ) deficient cells. The data reveal two DNA damage response pathways not previously observed upon deletion of individual shelterin proteins. The shelterin-free telomeres are processed by microhomology-mediated alternative-NHEJ when Ku70/80 is absent and are attacked by nucleolytic degradation in the absence of 53BP1. The data establish that the end-protection problem is specified by six pathways [ATM (ataxia telangiectasia mutated) and ATR (ataxia telangiectasia and Rad3 related) signaling, classical-NHEJ, alt-NHEJ, homologous recombination, and resection] and show how shelterin acts with general DNA damage response factors to solve this problem.
Figures
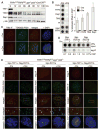
Fig. 1
Shelterin-free telomeres. (A) Immunoblots for TRF1, TRF2, and Rap1 after 4-OHT−induced TRF1/2 DKO from Lig4−/−p53−/−Cre-ERT2 MEFs. (B) ChIP for telomeric DNA associated with shelterin proteins in TRF1F/FTRF2F/Fp53−/−Lig4−/−MEFs (day 5 after H&R-Cre). Bars average percentage of telomeric DNA recovered in two independent experiments, ± SEMs. (C) IF-FISH for TIN2 at telomeres in TRF1F/FTRF2F/Fp53−/−Lig4−/−MEFs day 5 after H&R-Cre. TIN2 IF (red); telomeric PNA probe [fluorescein isothiocyanate (FITC), green]. (D) ChIP for telomeric DNA associated with Myc-TPP1, Myc-POT1a, and Flag-POT1b in TRF1F/FTRF2F/Fp53−/− Lig4−/−cells, with (+) and without (−) H&R-Cre. (E) IF for the telomeric localization of Myc-TPP1, Myc-POT1a, and Flag-POT1b (red, MYC or Flag antibodies) in TRF1F/FTRF2F/Fp53−/−Lig4−/− MEFs (5 days after H&R-Cre). Green, telomeric PNA probe or TRF1 IF.
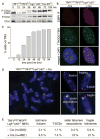
Fig. 2
Telomere dysfunction upon shelterin loss. (A) Induction of P-Chk1 and P-Chk2 after TRF1/2 codeletion. (B) IF-FISH assay for TIFs (telomere dysfunction-induced foci) in TRF1F/FTRF2F/FLig4−/−p53−/−Cre-ERT2 MEFs (5 days after Cre). FISH for telomeres (green), IF for 53BP1 (red), and 4′,6-diamidino-2-phenylindole (DAPI) as DNA counterstain (blue). (C) Time course of TIF response as in (B). TIFs were scored in TRF1F/FTRF2F/FLig4−/−p53−/−Cre-ERT2 cells at the indicated time points after 4-OHT. Cells with ≥5 telomeric 53BP1 foci were scored as TIF positive (n > 100 nuclei per time point). (D) Metaphase spread from TRF1F/FTRF2F/FLig4−/− p53−/− cells at 108 hours after Cre treatment, analyzed by telomeric CO-FISH using a FITC-OO-[CCCTAA]3 PNA probe (green) and a Tamra-OO-[TTAGGG]3 PNA probe (red). Blue, DAPI. Examples of fragile telomeres, chromosome- and chromatid-type fusions, sister telomere associations, and T-SCEs are on the right. (E) Quantification of aberrant telomeres in Cre-treated TRF1F/FTRF2F/FLig4−/−p53−/− MEFs analyzed as in (D).
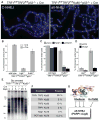
Fig. 3
Lig3- and PARP1-dependent alt-NHEJ in the absence of shelterin. (A) Metaphase chromosomes of the indicated MEFs analyzed (as in Fig. 2D) at 108 hours after Cre. (B) Quantification of telomere fusions in the indicated MEFs at 108 hours after H&R-Cre. Bars and not error bars means of three independent experiments, ± SDs. (C) Quantification of telomere fusions induced by deleting TRF1 and TRF2 [as in (A)] after treatment with PARP1 shRNA or 0.5 μM olaparib. (D) Quantification of telomere fusions [as in (C)] in cells treated with Lig3 or control shRNA. (E) Alt-NHEJ in G0 arrested TRF1F/FTRF2F/FKu80−/−p53+/+Cre-ERT2 MEFs. _Mbo_I and _Alu_I digested DNA resolved on a pulsed-field gel electrophoresis probed with end-labeled [AACCCT]4. Dashed and solid lines: fused and unfused telomeres, respectively. Day 4R: cells released on day 4 and analyzed on day 5. (F) Percentage of fused telomeres in Ku-deficient MEFs lacking the indicated shelterin subunit(s). Cells were analyzed at 108 hours after Cre-mediated deletion of the floxed alleles of shelterin. (G) Summary of the repression of Lig3- and PARP1-dependent alt-NHEJ by shelterin and Ku70/80.
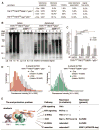
Fig. 4
53BP1 blocks 5′ end resection and shortening of shelterin-free telomeres. (A) Quantification of telomere aberrations in Cre-treated (108 hours) TRF1F/FTRF2F/F53BP1−/− p53−/− and TRF1F/FTRF2F/FLig4−/−p53−/−MEFs. *, 93% of the cells had ~12% of chromosome ends fused, whereas 7% of the cells had more than 50% of the chromosome ends fused. (B) Representative in-gel 3′ overhang analysis of the indicated MEFs after Cre treatment. Relative overhang signals were normalized to total telomeric DNA (lanes without Cre set to 1). (C) Quantification of 3′ overhangs of TRF1F/FTRF2F/F53BP1−/−p53−/− MEFs (+ or − H&R-Cre, 108 hours) treated with Exo1, CtIP, and Blm shRNAs. The ss/total signal ratios of the +Cre samples are expressed relative to the −Cre samples for each shRNA treatment. Means of three independent experiments ± SDs. P values: two-tailed student’s t tests. (D) Q-FISH of telomeres in TRF1F/F TRF2F/F53BP1−/−p53−/−MEFs with or without H&R-Cre (day 5). (E) Summary of the end-protection problem.
Similar articles
- No overt nucleosome eviction at deprotected telomeres.
Wu P, de Lange T. Wu P, et al. Mol Cell Biol. 2008 Sep;28(18):5724-35. doi: 10.1128/MCB.01764-07. Epub 2008 Jul 14. Mol Cell Biol. 2008. PMID: 18625717 Free PMC article. - 53BP1 deficiency combined with telomere dysfunction activates ATR-dependent DNA damage response.
Martínez P, Flores JM, Blasco MA. Martínez P, et al. J Cell Biol. 2012 Apr 16;197(2):283-300. doi: 10.1083/jcb.201110124. J Cell Biol. 2012. PMID: 22508511 Free PMC article. - BRCA1 and CtIP promote alternative non-homologous end-joining at uncapped telomeres.
Badie S, Carlos AR, Folio C, Okamoto K, Bouwman P, Jonkers J, Tarsounas M. Badie S, et al. EMBO J. 2015 Feb 3;34(3):410-24. doi: 10.15252/embj.201488947. Epub 2015 Jan 12. EMBO J. 2015. PMID: 25582120 Free PMC article. - Shelterin-Mediated Telomere Protection.
de Lange T. de Lange T. Annu Rev Genet. 2018 Nov 23;52:223-247. doi: 10.1146/annurev-genet-032918-021921. Epub 2018 Sep 12. Annu Rev Genet. 2018. PMID: 30208292 Review. - The Connection Between Cell Fate and Telomere.
Engin AB, Engin A. Engin AB, et al. Adv Exp Med Biol. 2021;1275:71-100. doi: 10.1007/978-3-030-49844-3_3. Adv Exp Med Biol. 2021. PMID: 33539012 Review.
Cited by
- Combatting cellular immortality in cancers by targeting the shelterin protein complex.
Chakraborty S, Banerjee S. Chakraborty S, et al. Biol Direct. 2024 Nov 22;19(1):120. doi: 10.1186/s13062-024-00552-4. Biol Direct. 2024. PMID: 39578854 Free PMC article. - Telomere length and cognitive changes in 7,877 older UK adults of European ancestry.
Packer A, Habiballa L, Tato-Barcia E, Breen G, Brooker H, Corbett A, Arathimos R, Ballard C, Hampshire A, Palmer A, Dima D, Aarsland D, Creese B, Malanchini M, Powell TR. Packer A, et al. Front Aging. 2024 Nov 1;5:1480326. doi: 10.3389/fragi.2024.1480326. eCollection 2024. Front Aging. 2024. PMID: 39553389 Free PMC article. - Is physical fitness associated with leucocyte telomere length in youth with type 1 diabetes?
Muñoz-Pardeza J, López-Gil JF, Huerta-Uribe N, Hormazábal-Aguayo I, Ojeda-Rodríguez A, Del Moral AM, Izquierdo M, García-Hermoso A. Muñoz-Pardeza J, et al. Pediatr Res. 2024 Nov 14. doi: 10.1038/s41390-024-03732-0. Online ahead of print. Pediatr Res. 2024. PMID: 39543404 - Telomere maintenance and the DNA damage response: a paradoxical alliance.
Harman A, Bryan TM. Harman A, et al. Front Cell Dev Biol. 2024 Oct 17;12:1472906. doi: 10.3389/fcell.2024.1472906. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39483338 Free PMC article. - Immune therapeutic strategies for the senescent tumor microenvironment.
Yasuda T, Alan Wang Y. Yasuda T, et al. Br J Cancer. 2024 Oct 28. doi: 10.1038/s41416-024-02865-7. Online ahead of print. Br J Cancer. 2024. PMID: 39468331 Review.
References
- Palm W, de Lange T. Annu Rev Genet. 2008;42:301. - PubMed
- Celli GB, de Lange T. Nat Cell Biol. 2005;7:712. - PubMed
- Celli GB, Denchi EL, de Lange T. Nat Cell Biol. 2006;8:885. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM049046/GM/NIGMS NIH HHS/United States
- AG016642/AG/NIA NIH HHS/United States
- R56 AG016642/AG/NIA NIH HHS/United States
- R37 GM049046/GM/NIGMS NIH HHS/United States
- R01 AG016642/AG/NIA NIH HHS/United States
- R01 CA076027/CA/NCI NIH HHS/United States
- GM49046/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous