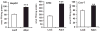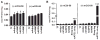Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase - PubMed (original) (raw)
. 2012 May 2;15(5):691-702.
doi: 10.1016/j.cmet.2012.04.008.
Gabriele Schoiswohl, Chandramohan Chitraju, Margret Paar, Irina Cornaciu, Ashraf Y Rangrez, Nuttaporn Wongsiriroj, Harald M Nagy, Pavlina T Ivanova, Sarah A Scott, Oskar Knittelfelder, Gerald N Rechberger, Ruth Birner-Gruenberger, Sandra Eder, H Alex Brown, Guenter Haemmerle, Monika Oberer, Achim Lass, Erin E Kershaw, Robert Zimmermann, Rudolf Zechner
Affiliations
- PMID: 22560221
- PMCID: PMC3361708
- DOI: 10.1016/j.cmet.2012.04.008
Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase
Manju Kumari et al. Cell Metab. 2012.
Abstract
Numerous studies in humans link a nonsynonymous genetic polymorphism (I148M) in adiponutrin (ADPN) to various forms of fatty liver disease and liver cirrhosis. Despite its high clinical relevance, the molecular function of ADPN and the mechanism by which I148M variant affects hepatic metabolism are unclear. Here we show that ADPN promotes cellular lipid synthesis by converting lysophosphatidic acid (LPA) into phosphatidic acid. The ADPN-catalyzed LPA acyltransferase (LPAAT) reaction is specific for LPA and long-chain acyl-CoAs. Wild-type mice receiving a high-sucrose diet exhibit substantial upregulation of Adpn in the liver and a concomitant increase in LPAAT activity. In Adpn-deficient mice, this diet-induced increase in hepatic LPAAT activity is reduced. Notably, the I148M variant of human ADPN exhibits increased LPAAT activity leading to increased cellular lipid accumulation. This gain of function provides a plausible biochemical mechanism for the development of liver steatosis in subjects carrying the I148M variant.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Figure 1. Overexpression of Adpn Results in Increased TG Synthesis
Accumulation of [1-14C] oleic acid into TGs was assessed in HepG2, CHO, and Cos-7 cells. HepG2 and CHO cells were infected with murine Adpn expressing adenovirus, and Cos-7 cells were transfected with an expression vector encoding murine Adpn. Adenovirus or vector DNA encoding β-galactosidase (LacZ) was used as a control. After 36 hr of infection or transfection, cells were incubated with 1 μCi/ml [1-14C] oleic acid and 400 μM oleic acid complexed to BSA for 6 hr. Subsequently, lipids were extracted and separated by TLC. Radioactivity comigrating with the TG band was excised and quantified by scintillation counting. Data are presented as mean ± SD and represent two independent experiments with each sample in triplicate. Statistical significance was determined by a two-tailed Student’s t test, **p < 0.01, ***p < 0.001.
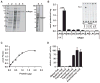
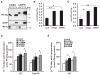
Figure 2. Recombinant mAdpn Possesses Lysophosphatidic Acid Acyltransferase Activity
(A) Purification of recombinant murine Adpn (mAdpn) and TF by metal-ion affinity chromatography. Lane 1, protein standard; lane 2, uninduced E. coli lysate; lane 3, IPTG-induced lysate (E. coli BL21 pCold-mAdpn); lane 4, flowthrough of Ni-NTA column; lane 5, eluate of Ni-NTA column containing mAdpn; lane 6, IPTG-induced lysate (E. coli BL21 pCold); lane 7, eluate of Ni-NTA column containing TF. (B) Purified mAdpn or TF (1 μg protein) was incubated with 20 μM [1-14C] oleoyl-CoA as acyl donor in the presence of 100 μM of various acyl acceptors (lysophosphatidic acid [LPA], MG, DG, glycerol-3-phosphate [G3P], lysophosphatidyl choline [LPC], lysophosphatidyl ethanolamine [LPE], lysophosphatidyl inositol [LPI], or lysophosphatidyl serine [LPS]) for 10 min at 37°C. Lipids were solvent extracted and separated by TLC. Enzymatic activity was calculated from the radioactivity associated with the product band. The inset represents a phosphorimager scan of the TLC plate showing that mAdpn is active only in the presence of oleoyl-CoA and LPA but not with other lysophospholipids. PA, phosphatidic acid. Purified TF incubated with oleoyl-CoA and LPA was used as a negative control (represented as TF in the inset). LPAAT activity of purified mAdpn was measured (C) at various protein concentrations and (D) in the presence of acyl-CoAs with varying fatty acid chain length. Relative LPAAT activity was determined by normalizing the specific activity of mAdpn (2.4 nmol/min*mg) in presence of [3H] LPA and oleoyl-CoA to 100%. Data are presented as mean ± SD and are representative of three independent experiments with each sample in duplicate. Statistical significance was determined by a two-tailed Student’s t test, ***p < 0.001.
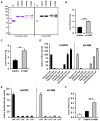
Figure 3. LPAAT Activity of Purified Murine and Human I148M Variants
(A) Coomassie blue stain showing purified TF and recombinant mAdpn, mI148M, hADPN, and hI148M. I148M mutants were generated by site-directed mutagenesis, purified, and analyzed for LPAAT activity in comparison to wild-type proteins. Purified proteins were detected by western blotting using anti-His monoclonal antibody. (B and C) (B) LPAAT activity of mAdpn and mI148M and (C) hADPN and hI148M using 1 μg of purified protein in a reaction with 20 μM [1-14C] oleoyl-CoA and 100 μM LPA. Purified TF was used as negative control with negligible LPAAT activity (<0.1 nmol PA/min*mg). (D) Substrate specificity of recombinant hADPN and hI148M for acyl donors was measured by incubating 2 μg of partially purified proteins with 25 μM [3H] LPA (600,000 cpm/assay) and 20 μM of various acyl-CoAs. Relative LPAAT activity was determined by normalizing the specific activity of hADPN and hI148M (4.2 and 8.1 nmol/min*mg, respectively) in the presence of [3H] LPA and oleoyl-CoA to 100%. (E) Substrate specificity of recombinant hADPN and hI148M for acyl acceptors was measured by incubating 2 μg of partially purified proteins with 20 μM [1-14C] oleoyl-CoA as acyl donor in the presence of 100 μM of various acyl acceptors (LPA, lysophosphatidyl choline [LPC], lysophosphatidyl ethanolamine [LPE], lysophosphatidyl inositol [LPI], or lysophosphatidyl serine [LPS]). Relative LPAAT activity was determined by normalizing the specific activity of hADPN and hI148M (as shown in Figure 3C) in the presence of LPA and [1-14C] oleoyl-CoA to 100%. (F) Acylation of glycerol-3-phosphate (G3P) was measured by incubating 2 μg purified TF, hADPN, and hI148M with 300 μM G3P and 20 μM [1-14C] oleoyl-CoA. Data are presented as mean ± SD and are representative of at least two independent experiments with each sample in duplicates. Statistical significance was determined by a two-tailed Student’s t test, **p < 0.01, ***p < 0.001.
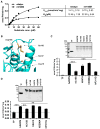
Figure 4. Enzyme Properties of Murine and Human ADPN Variants
(A) LPAAT activity of mAdpn and mI148M was monitored as a function of [1-14C] oleoyl-CoA concentration and kinetic parameters were calculated by Michaelis-Menten plot using GraphPad Prism software. (B) Structure around the putative “catalytic dyad” (Ser-47 and Asp-166) involved in the proposed lipase activity of ADPN. The structure was deduced from an alignment of Leu10-Phe175 of ADPN to Leu32-Ala225 of Pat17 (PDB ID 1OWX), the best-scoring template with known 3D structure using the Phyre2 server (Kelley and Sternberg, 2009), and PyMOL (The PyMOL Molecular Graphics System, Version 1.4.1, Schrödinger, LLC). The model reveals close spatial proximity of Ile148 (highlighted in yellow) and Cys15 (highlighted in green) to the proposed active site (highlighted in red). (C) LPAAT activity of hADPN and hADPN mutants with point mutations at Cys-15 to Ser (hC15S), Ser-47 to Ala (hS47A), Asp-206 to Ala (hD206A), and Pro-311 to Gly (hP311G). Mutants were generated by site-directed mutagenesis and purified under conditions identical to those of the wild-type protein. The inset shows a Coomassie blue-stained SDS-PAGE gel with various purified recombinant ADPN variants (2 μg protein/lane). LPAAT activity was measured using 2 μg of purified proteins or TF as described in the Experimental Procedures. (D) LPAAT activity was determined in bacterial total lysates (10 μg lysate protein) overexpressing TF, hADPN, or hADPN mutants. Protein overexpression was confirmed by western blotting in bacterial total lysates using anti-His monoclonal antibody (inset). Experiments were done in duplicates and repeated twice. Data are presented as mean ± SD. Statistical significance was determined by a two-tailed Student’s t test, **p < 0.01, ***p < 0.001.
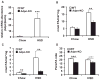
Figure 5. TG Hydrolase Activity of ADPN
(A) TG hydrolase activity assay using radiolabeled triolein as substrate in the presence of 5 μg purified TF, mAdpn, mI148M, hADPN, hI48M, and (B) bacterial total lysates (40 μg lysate protein) overexpressing TF, mAdpn, mI148M, hADPN, hI148M, and strep-tagged mATGL. Assays were performed in the absence or presence of purified mCGI-58. Data are shown as mean ± SD and represent three independent experiments. Statistical significance was determined by a two-tailed Student’s t test, *p < 0.05, ***p < 0.001. FFA, free fatty acid.
Figure 6. Overexpression of ADPN in Mammalian Cells Increases LPAAT Activity and Lipid Synthesis
Cos-7 cells were transfected with His-tagged pcDNA4/HisMax expression vector encoding mouse or human ADPN and LacZ as a control. After 24 hr, cells were loaded with 400 μM oleic acid complexed to FA-free BSA (3:1) in DMEM containing 10% FBS for 20 hr. LDs, membrane, and cytosol fractions were isolated using density gradient centrifugation. (A) Expression of recombinant proteins was determined by immunoblotting using anti-His monoclonal antibody. Marker proteins for LDs and total membrane fractions were detected by anti-ADRP and anti-IRE1α antibody, respectively. (B and C) (B) LPAAT activity assay using 20 μM [1-14C] oleoyl-CoA as acyl donor and 100 μM LPA as acyl acceptor in total membrane fractions (2 μg of total membrane protein) and (C) isolated LD fractions (20 μg of LD protein). Experiments were done in triplicate and repeated twice. Data are shown as mean ± SD. Statistical significance was determined by a two-tailed Student’s t test, *p < 0.05 and **p < 0.01. (D and E) (D) Incorporation of [1-14C] oleic acid into TGs and total PLs in Cos-7 cells overexpressing mouse or human ADPN or their respective I148M mutant proteins, in the presence of high-glucose medium. Lipids were separated by TLC and radioactivity comigrating with TGs and total PLs, and (E) phosphatidylcholine (PC) was quantified by liquid scintillation counting. Experiments were done in duplicate and repeated thrice. The graph depicts the mean ± SD of two experiments. Statistical significance was determined by a two-tailed Student’s t test, *p < 0.05 (compared to LacZ), #p < 0.05 (compared to mouse/human WT ADPN).
Figure 7. Hepatic LPAAT Activity of Wild-Type and Adiponutrin-Deficient Mice
Five hours postfeeding, male wild-type (WT) and _Adpn_-KO mice fed a chow diet or HSD were sacrificed, and the liver was excised. (A) Relative liver Adpn mRNA expression as determined by RT-qPCR was normalized to β-actin as a reference gene and compared to WT mice fed chow diet, n = 6. (B) LPAAT activity was determined in liver tissue homogenates (20 μg total protein), n = 5. (C) LD-associated LPAAT activity was determined in freshly prepared liver LDs (10 μg LD protein). Data are representative of three independent diet studies, n = 4 or 5. Data are shown as mean ± SD. Statistical significance was determined by a two-tailed student’s t test, **p < 0.01, ***p < 0.001. (D) Phosphatidic acid (PA) to lysophosphatidic acid (LPA) ratio in WT and _Adpn_-KO mice liver lipid extracts. Liver samples were collected from chow diet- and HSD-fed WT and _Adpn_-KO mice after 2 min organ perfusion with Krebs-Henseleit Buffer 2. For the determination of LPA and PA concentrations, hepatic glycerophospholipids were extracted using a modified Bligh and Dyer method (Bligh and Dyer, 1959). Lipids were quantified by LC/MS/MS (details described in the Supplemental Experimental Procedures), n = 6. Data are shown as mean ± SEM. Statistical significance was determined by post hoc analysis, ***p < 0.001.
Similar articles
- ATP-independent fatty acyl-coenzyme A synthesis from phospholipid: coenzyme A-dependent transacylation activity toward lysophosphatidic acid catalyzed by acyl-coenzyme A:lysophosphatidic acid acyltransferase.
Yamashita A, Kawagishi N, Miyashita T, Nagatsuka T, Sugiura T, Kume K, Shimizu T, Waku K. Yamashita A, et al. J Biol Chem. 2001 Jul 20;276(29):26745-52. doi: 10.1074/jbc.M101795200. Epub 2001 May 14. J Biol Chem. 2001. PMID: 11352914 - Identification of an intrinsic lysophosphatidic acid acyltransferase activity in the lipolytic inhibitor G0/G1 switch gene 2 (G0S2).
Zhang X, Xie X, Heckmann BL, Saarinen AM, Gu H, Zechner R, Liu J. Zhang X, et al. FASEB J. 2019 May;33(5):6655-6666. doi: 10.1096/fj.201802502R. Epub 2019 Feb 25. FASEB J. 2019. PMID: 30802154 Free PMC article. - SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast.
Benghezal M, Roubaty C, Veepuri V, Knudsen J, Conzelmann A. Benghezal M, et al. J Biol Chem. 2007 Oct 19;282(42):30845-55. doi: 10.1074/jbc.M702719200. Epub 2007 Aug 3. J Biol Chem. 2007. PMID: 17675291 - The role of PNPLA3 in health and disease.
Pingitore P, Romeo S. Pingitore P, et al. Biochim Biophys Acta Mol Cell Biol Lipids. 2019 Jun;1864(6):900-906. doi: 10.1016/j.bbalip.2018.06.018. Epub 2018 Jun 20. Biochim Biophys Acta Mol Cell Biol Lipids. 2019. PMID: 29935383 Review. - Acyltransferases and transacylases involved in fatty acid remodeling of phospholipids and metabolism of bioactive lipids in mammalian cells.
Yamashita A, Sugiura T, Waku K. Yamashita A, et al. J Biochem. 1997 Jul;122(1):1-16. doi: 10.1093/oxfordjournals.jbchem.a021715. J Biochem. 1997. PMID: 9276665 Review.
Cited by
- Omic studies reveal the pathogenic lipid droplet proteins in non-alcoholic fatty liver disease.
Zhang X, Wang Y, Liu P. Zhang X, et al. Protein Cell. 2017 Jan;8(1):4-13. doi: 10.1007/s13238-016-0327-9. Epub 2016 Oct 18. Protein Cell. 2017. PMID: 27757845 Free PMC article. Review. - Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis.
Li JZ, Huang Y, Karaman R, Ivanova PT, Brown HA, Roddy T, Castro-Perez J, Cohen JC, Hobbs HH. Li JZ, et al. J Clin Invest. 2012 Nov;122(11):4130-44. doi: 10.1172/JCI65179. J Clin Invest. 2012. PMID: 23023705 Free PMC article. - Genetic Factors in the Pathogenesis of Nonalcoholic Fatty Liver and Steatohepatitis.
Dongiovanni P, Romeo S, Valenti L. Dongiovanni P, et al. Biomed Res Int. 2015;2015:460190. doi: 10.1155/2015/460190. Epub 2015 Jul 27. Biomed Res Int. 2015. PMID: 26273621 Free PMC article. Review. - The Role of Macronutrients in the Pathogenesis, Prevention and Treatment of Non-Alcoholic Fatty Liver Disease (NAFLD) in the Paediatric Population-A Review.
Pixner T, Stummer N, Schneider AM, Lukas A, Gramlinger K, Julian V, Thivel D, Mörwald K, Maruszczak K, Mangge H, Gomahr J, Weghuber D, Furthner D. Pixner T, et al. Life (Basel). 2022 Jun 5;12(6):839. doi: 10.3390/life12060839. Life (Basel). 2022. PMID: 35743870 Free PMC article. Review. - PNPLA3, CGI-58, and Inhibition of Hepatic Triglyceride Hydrolysis in Mice.
Wang Y, Kory N, BasuRay S, Cohen JC, Hobbs HH. Wang Y, et al. Hepatology. 2019 Jun;69(6):2427-2441. doi: 10.1002/hep.30583. Epub 2019 Apr 9. Hepatology. 2019. PMID: 30802989 Free PMC article.
References
- Agarwal AK, Arioglu E, De Almeida S, Akkoc N, Taylor SI, Bowcock AM, Barnes RI, Garg A. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat Genet. 2002;31:21–23. - PubMed
- Baulande S, Lasnier F, Lucas M, Pairault J. Adiponutrin, a transmembrane protein corresponding to a novel dietary- and obesity-linked mRNA specifically expressed in the adipose lineage. J Biol Chem. 2001;276:33336–33344. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 GM069338-10/GM/NIGMS NIH HHS/United States
- U54 GM069338/GM/NIGMS NIH HHS/United States
- F 3016/FWF_/Austrian Science Fund FWF/Austria
- Z 136/FWF_/Austrian Science Fund FWF/Austria
- HHMI/Howard Hughes Medical Institute/United States
- P30 DK036836/DK/NIDDK NIH HHS/United States
- P 22170/FWF_/Austrian Science Fund FWF/Austria
- F 3002/FWF_/Austrian Science Fund FWF/Austria
- P30 DK-036836/DK/NIDDK NIH HHS/United States
- F 3001/FWF_/Austrian Science Fund FWF/Austria
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous