The updated biology of hypoxia-inducible factor - PubMed (original) (raw)
Review
The updated biology of hypoxia-inducible factor
Samantha N Greer et al. EMBO J. 2012.
Abstract
Oxygen is essential for eukaryotic life and is inextricably linked to the evolution of multicellular organisms. Proper cellular response to changes in oxygen tension during normal development or pathological processes, such as cardiovascular disease and cancer, is ultimately regulated by the transcription factor, hypoxia-inducible factor (HIF). Over the past decade, unprecedented molecular insight has been gained into the mammalian oxygen-sensing pathway involving the canonical oxygen-dependent prolyl-hydroxylase domain-containing enzyme (PHD)-von Hippel-Lindau tumour suppressor protein (pVHL) axis and its connection to cellular metabolism. Here we review recent notable advances in the field of hypoxia that have shaped a more complex model of HIF regulation and revealed unique roles of HIF in a diverse range of biological processes, including immunity, development and stem cell biology.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
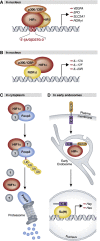
Expanded model of canonical HIFα regulation. (A) Under normal oxygen tension, HIFα is subject to oxygen-dependent prolyl hydroxylation by PHDs, which allows for substrate recognition and ubiquitylation by pVHL and its associated ubiquitin–ligase complex. Polyubiquitylated HIFα is degraded by the 26S proteasome. The prolyl-hydroxylase activity of PHDs is regulated by a number of intracellular factors, including ROS, which are in turn negatively modulated by SIRT3. Binding of the HIFα coactivator p300/CBP is inhibited by asparaginyl hydroxylation by FIH. HIFα is upregulated at the mRNA level by mTOR and STAT3, while SIRT6 negatively regulates HIFα protein levels. (B) Under low oxygen tension HIFα escapes prolyl hydroxylation by PHDs and associates with nuclear HIFβ. The heterodimer binds to a core consensus sequence at the promoters of HIF-responsive genes, and upon binding to the coactivators p300/CBP and PKM2, initiates transcription. The interaction between HIFα and p300 may be regulated by a variety of factors that sterically impede binding or add/remove PTMs to influence the transcriptional activity of HIFα. See text for details (PHD, prolyl-hydroxylase domain-containing enzyme; NO, nitric oxide; SIRT1/3/6, sirtuin 1/3/6; FIH, factor inhibiting HIF; CBP, Creb-binding protein; OH, hydroxyl group; mTOR, mammalian target of rapamycin; STAT3, signal transducer and activator of transcription 3; ub, ubiquitin moiety; EloB/C, elongins B and C; Cul2, cullin 2; Rbx 1, RING-box protein 1; pVHL, von Hippel-Lindau protein; ROS, reactive oxygen species; HIF, hypoxia-inducible factor; CITED2/4, CBP/p300 interacting transactivator with ED-rich tail 2/4; PCAF, p300/CBP-associated factor; SENP1/3, sentrin-specific protease 1/3; PKM2, pyruvate kinase isoform M2; hnRNPs, heterogeneous nuclear ribonucleoproteins).
Figure 2
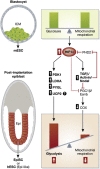
HIF1α regulates the metabolic and phenotypic transition from ESC to EpiSC. mESC isolated from the inner cell mass (ICM) of the blastocyst are metabolically and phenotypically distinct from post-implantation EpiSC isolated from the epiblast (Epi) or hESC (Epi-like). HIF1α expression regulates the metabolic and phenotypic transition from ESC to EpiSC. Increased HIF1α expression in EpiSC/hESC promotes a highly glycolytic metabolism via increased expression of glycolytic genes such as PDK1, LDHA, and PYGL and downregulation of mitochondrial COX gene expression via TGFβ/Activin/Nodal signalling.
Figure 3
Canonical and non-canonical regulation by HIFα. (A) Classical DNA-binding transcription factor: HIFα associates with HIFβ and transcriptional coactivators in the nucleus, binds conserved hypoxia-response elements in the DNA sequence of target promoters, and initiates transcription of HIF-target genes. (B) Transcriptional coactivator: HIF1α binds RORδt, but not DNA, at the promoters of RORδt-responsive genes in the nucleus. Recruitment of p300 by HIF1α leads to deacetylation of histones H3 and H4 thereby facilitating transcription. (C) Director for proteasome-mediated degradation: HIF1α binds to Foxp3, presumably in the cytoplasm, and in association with an unidentified degradation complex mediates its ubiquitylation and subsequent proteasomal degradation. (D) Stabilizing force for endocytosed receptors: HIFα (Sima) colocalizes with and stabilizes full-length Notch that has been internalized from the plasma membrane in early endosomes. See text for details (HIF, hypoxia-inducible factor; CBP, Creb-binding protein; IL, interleukin; ub, ubiquitin moiety; ICD, intracellular domain; Su(H), suppressor of Hairless).
Similar articles
- Regulation of HIF by the von Hippel-Lindau tumour suppressor: implications for cellular oxygen sensing.
Mole DR, Maxwell PH, Pugh CW, Ratcliffe PJ. Mole DR, et al. IUBMB Life. 2001 Jul;52(1-2):43-7. doi: 10.1080/15216540252774757. IUBMB Life. 2001. PMID: 11795592 Review. - Transgenic models to understand hypoxia-inducible factor function.
Doedens A, Johnson RS. Doedens A, et al. Methods Enzymol. 2007;435:87-105. doi: 10.1016/S0076-6879(07)35005-2. Methods Enzymol. 2007. PMID: 17998050 Review. - Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein.
Cockman ME, Masson N, Mole DR, Jaakkola P, Chang GW, Clifford SC, Maher ER, Pugh CW, Ratcliffe PJ, Maxwell PH. Cockman ME, et al. J Biol Chem. 2000 Aug 18;275(33):25733-41. doi: 10.1074/jbc.M002740200. J Biol Chem. 2000. PMID: 10823831 - Kelch-like 20 up-regulates the expression of hypoxia-inducible factor-2α through hypoxia- and von Hippel-Lindau tumor suppressor protein-independent regulatory mechanisms.
Higashimura Y, Terai T, Yamaji R, Mitani T, Ogawa M, Harada N, Inui H, Nakano Y. Higashimura Y, et al. Biochem Biophys Res Commun. 2011 Sep 23;413(2):201-5. doi: 10.1016/j.bbrc.2011.08.058. Epub 2011 Aug 24. Biochem Biophys Res Commun. 2011. PMID: 21888897 - Hypoxia-inducible factor 1 (HIF-1) pathway.
Semenza GL. Semenza GL. Sci STKE. 2007 Oct 9;2007(407):cm8. doi: 10.1126/stke.4072007cm8. Sci STKE. 2007. PMID: 17925579 Review.
Cited by
- Genetic Signatures of Positive Selection in Human Populations Adapted to High Altitude in Papua New Guinea.
González-Buenfil R, Vieyra-Sánchez S, Quinto-Cortés CD, Oppenheimer SJ, Pomat W, Laman M, Cervantes-Hernández MC, Barberena-Jonas C, Auckland K, Allen A, Allen S, Phipps ME, Huerta-Sanchez E, Ioannidis AG, Mentzer AJ, Moreno-Estrada A. González-Buenfil R, et al. Genome Biol Evol. 2024 Aug 5;16(8):evae161. doi: 10.1093/gbe/evae161. Genome Biol Evol. 2024. PMID: 39173139 Free PMC article. - Metabolic theory of preeclampsia: implications for maternal cardiovascular health.
Manoharan MM, Montes GC, Acquarone M, Swan KF, Pridjian GC, Nogueira Alencar AK, Bayer CL. Manoharan MM, et al. Am J Physiol Heart Circ Physiol. 2024 Sep 1;327(3):H582-H597. doi: 10.1152/ajpheart.00170.2024. Epub 2024 Jul 5. Am J Physiol Heart Circ Physiol. 2024. PMID: 38968164 Review. - Differential impact of sex on regulation of skeletal muscle mitochondrial function and protein homeostasis by hypoxia-inducible factor-1α in normoxia.
Welch N, Mishra S, Bellar A, Kannan P, Gopan A, Goudarzi M, King J, Luknis M, Musich R, Agrawal V, Bena J, Koch CJ, Li L, Willard B, Shah YM, Dasarathy S. Welch N, et al. J Physiol. 2024 Jun;602(12):2763-2806. doi: 10.1113/JP285339. Epub 2024 May 18. J Physiol. 2024. PMID: 38761133 - Prognostic biomarker HIF1α and its correlation with immune infiltration in gliomas.
Ding Z, Zhang J, Li L, Wang C, Mei J. Ding Z, et al. Oncol Lett. 2024 Mar 4;27(5):193. doi: 10.3892/ol.2024.14326. eCollection 2024 May. Oncol Lett. 2024. PMID: 38495835 Free PMC article. - Understanding the molecular mechanism responsible for developing therapeutic radiation-induced radioresistance of rectal cancer and improving the clinical outcomes of radiotherapy - A review.
Jain SM, Nagainallur Ravichandran S, Murali Kumar M, Banerjee A, Sun-Zhang A, Zhang H, Pathak R, Sun XF, Pathak S. Jain SM, et al. Cancer Biol Ther. 2024 Dec 31;25(1):2317999. doi: 10.1080/15384047.2024.2317999. Epub 2024 Mar 6. Cancer Biol Ther. 2024. PMID: 38445632 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous