Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium - PubMed (original) (raw)
Editorial
Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium
Steve D M Brown et al. Dis Model Mech. 2012 May.
No abstract available
Figures
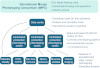
Fig. 1.
Organisation of the IMPC. A core of mouse production and phenotyping centres is allied to the data centre supporting the IMPC. The core mouse production and phenotyping centres also network with a variety of other biomedical research centres that will undertake more specialised phenotyping on selected lines, as well as provide additional expertise and input into the IMPC phenotyping programme. COPD, chronic obstructive pulmonary disease; CVD, cardiovascular disease; Dev biol, developmental biology; KO, knockout; Neuro, neurological; QC, quality control.
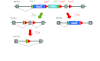
Fig. 2.
Design of the knockout first, conditional ready allele employed by the IMPC. The tm1a allele is generated in ES cells (which itself provides a knockout through splicing into the lacZ reporter). Mice generated with the tm1a allele are crossed to an appropriate Cre driver line to both eliminate a key exon within the gene and remove the selection cassette, generating the tm1b null allele. The tm1b allele will be phenotyped by the IMPC. Alternatively, Flp action on the tm1a allele can produce the tm1c ‘conditional ready’ allele. Grey boxes represent exons, and grey lines show introns. Figure reproduced from Skarnes et al. (Skarnes et al., 2011) with permission.
Fig. 3.
The IMPC adult phenotyping pipeline. The consensus IMPC pipeline for phenotyping in the adult is shown, highlighting mandatory tests, non-mandatory tests (which a number of centres will employ, but not all) and tests under development. In-life phenotyping is carried out from 9 to 15 weeks, after which a variety of terminal tests are employed. ECG, electrocardiogram; Echo, echocardiogram; F, female; M, male; PPI, prepulse inhibition. Schematic reproduced from
, with permission.
Similar articles
- Commentary: The International Mouse Phenotyping Consortium: high-throughput in vivo functional annotation of the mammalian genome.
Lloyd KCK. Lloyd KCK. Mamm Genome. 2024 Dec;35(4):537-543. doi: 10.1007/s00335-024-10068-x. Epub 2024 Sep 10. Mamm Genome. 2024. PMID: 39254744 Free PMC article. - Bloomsbury report on mouse embryo phenotyping: recommendations from the IMPC workshop on embryonic lethal screening.
Adams D, Baldock R, Bhattacharya S, Copp AJ, Dickinson M, Greene ND, Henkelman M, Justice M, Mohun T, Murray SA, Pauws E, Raess M, Rossant J, Weaver T, West D. Adams D, et al. Dis Model Mech. 2013 May;6(3):571-9. doi: 10.1242/dmm.011833. Epub 2013 Mar 18. Dis Model Mech. 2013. PMID: 23519032 Free PMC article. - The International Mouse Phenotyping Consortium: comprehensive knockout phenotyping underpinning the study of human disease.
Groza T, Gomez FL, Mashhadi HH, Muñoz-Fuentes V, Gunes O, Wilson R, Cacheiro P, Frost A, Keskivali-Bond P, Vardal B, McCoy A, Cheng TK, Santos L, Wells S, Smedley D, Mallon AM, Parkinson H. Groza T, et al. Nucleic Acids Res. 2023 Jan 6;51(D1):D1038-D1045. doi: 10.1093/nar/gkac972. Nucleic Acids Res. 2023. PMID: 36305825 Free PMC article. - Mouse genetic and phenotypic resources for human genetics.
Schofield PN, Hoehndorf R, Gkoutos GV. Schofield PN, et al. Hum Mutat. 2012 May;33(5):826-36. doi: 10.1002/humu.22077. Hum Mutat. 2012. PMID: 22422677 Free PMC article. Review. - New models for human disease from the International Mouse Phenotyping Consortium.
Cacheiro P, Haendel MA, Smedley D; International Mouse Phenotyping Consortium and the Monarch Initiative. Cacheiro P, et al. Mamm Genome. 2019 Jun;30(5-6):143-150. doi: 10.1007/s00335-019-09804-5. Epub 2019 May 24. Mamm Genome. 2019. PMID: 31127358 Free PMC article. Review.
Cited by
- Controlling complexity: the clinical relevance of mouse complex genetics.
Schughart K, Libert C; SYSGENET consortium; Kas MJ. Schughart K, et al. Eur J Hum Genet. 2013 Nov;21(11):1191-6. doi: 10.1038/ejhg.2013.79. Epub 2013 May 1. Eur J Hum Genet. 2013. PMID: 23632795 Free PMC article. Review. - Use and misuse of material transfer agreements: lessons in proportionality from research, repositories, and litigation.
Bubela T, Guebert J, Mishra A. Bubela T, et al. PLoS Biol. 2015 Feb 3;13(2):e1002060. doi: 10.1371/journal.pbio.1002060. eCollection 2015 Feb. PLoS Biol. 2015. PMID: 25646804 Free PMC article. - From mice to humans.
McMurray F, Moir L, Cox RD. McMurray F, et al. Curr Diab Rep. 2012 Dec;12(6):651-8. doi: 10.1007/s11892-012-0323-2. Curr Diab Rep. 2012. PMID: 22996130 Free PMC article. Review. - Blastocyst genotyping for quality control of mouse mutant archives: an ethical and economical approach.
Scavizzi F, Ryder E, Newman S, Raspa M, Gleeson D, Wardle-Jones H, Montoliu L, Fernandez A, Dessain ML, Larrigaldie V, Khorshidi Z, Vuolteenaho R, Soininen R, André P, Jacquot S, Hong Y, de Angelis MH, Ramirez-Solis R, Doe B. Scavizzi F, et al. Transgenic Res. 2015 Oct;24(5):921-7. doi: 10.1007/s11248-015-9897-1. Epub 2015 Jul 16. Transgenic Res. 2015. PMID: 26178246 Free PMC article. - Histopathology reveals correlative and unique phenotypes in a high-throughput mouse phenotyping screen.
Adissu HA, Estabel J, Sunter D, Tuck E, Hooks Y, Carragher DM, Clarke K, Karp NA; Sanger Mouse Genetics Project; Newbigging S, Jones N, Morikawa L, White JK, McKerlie C. Adissu HA, et al. Dis Model Mech. 2014 May;7(5):515-24. doi: 10.1242/dmm.015263. Epub 2014 Mar 20. Dis Model Mech. 2014. PMID: 24652767 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources