The role of insulin-regulated aminopeptidase in MHC class I antigen presentation - PubMed (original) (raw)
The role of insulin-regulated aminopeptidase in MHC class I antigen presentation
Loredana Saveanu et al. Front Immunol. 2012.
Abstract
Production of MHC-I ligands from antigenic proteins generally requires multiple proteolytic events. While the proteolytic steps required for antigen processing in the endogenous pathway are clearly established, persisting gaps of knowledge regarding putative cross-presentation compartments have made it difficult to map the precise proteolytic events required for generation of cross-presented antigens. It is only in the past decade that the importance of aminoterminal trimming as the final step in the endogenous presentation pathway has been recognized and that the corresponding enzymes have been described. This review focuses on the aminoterminal trimming of exogenous cross-presented peptides, with particular emphasis on the identification of insulin responsive aminopeptidase (IRAP) as the principal trimming aminopeptidase in endosomes and phagosomes.
Keywords: ERAP; IRAP; MHC class I; aminopeptidase; cross-presentation; dendritic cells.
Figures
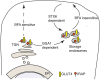
Figure 1
IRAP storage vesicles are slow recycling endosomes. Newly synthetized IRAP accumulates in storage endosomes as early as 3 h after transfection (Watson et al., 2004) and acquires insulin responsiveness after 6–9 h. During the first 3 h after enzyme synthesis, IRAP sorting from the ER is brefeldin A sensitive. Once the enzyme reaches the storage endosomes its translocation to the plasma membrane becomes insensitive to brefeldin A treatment (Watson et al., 2008). Sorting of newly synthetized IRAP from the TGN to the storage endosomes in adipocytes requires the GGA1 clathrin adaptor (Liu et al., ; Hou et al., 2006) and the LL76,77 sequence in the cytosolic domain of IRAP (Watson et al., 2008).
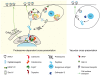
Figure 2
Cell-specific regulation of IRAP storage vesicles. (A) In insulin-responsive tissues, such as adipocytes and muscles, IRAP trafficking is regulated by insulin. Upon insulin binding to its receptor, PI3K-PDK1-Akt protein kinases are activated in a cascade. The most important effector in these cells seems to be the RabGAP AS160, which is phosphorylated by Akt2. Phosphorylation of AS160 leads to its dissociation from the storage endosomes and activation of Rab8, Rab10, and Rab14 that drive the endosome translocation to the cell surface. The biological effect is the increase at cell surface of the principal vesicle cargo, the glucose transporter GLUT4. (B) In other cells, the stimuli that regulate IRAP endosome trafficking and the signaling pathways involved in this process are poorly characterized. In DCs, IRAP endosomes are recruited rapidly to the phagosomal membrane. The phagocytic receptors and the signaling molecules responsible for this phenomenon are not yet identified.
Figure 3
Cross-presentation pathways. According to the involvement of the proteasome in antigen processing cross-presentation is divided in two main pathways: proteasome-dependent cross-presentation (left panel) and vacuolar cross-presentation (right panel). The vacuolar cross-presentation does not require proteasome activity; the entire antigen processing and MHC class I loading with cross-presented peptides occur inside the vacuole. The proteasome-dependent cross-presentation involves the transport of the exogenous antigens (possibly by Sec61) into the DC cytosol of the DC and generation of N-terminal extended precursors of MHC-I ligands in the cytosol. The aminoterminal trimming of these peptide precursors can occur in two different compartments: (i) in a cytosol to ER pathway, epitope precursors will join the endogenous processing pathway after their transport by TAP into the ER (Kovacsovics-Bankowski and Rock, 1995). In this case peptide trimming is carried out by ERAPs; (ii) in a cytosol to endosome pathway, the precursors of class I ligands are retro-transported into specialized endosomes (Burgdorf et al., ; Saveanu et al., 2009) or into phagosomes (Guermonprez et al., 2003) and final trimming is performed by IRAP (Saveanu et al., 2009).
Similar articles
- Insulin-regulated aminopeptidase and its compartment in dendritic cells.
Saveanu L, Babdor J, Lawand M, van Endert P. Saveanu L, et al. Mol Immunol. 2013 Sep;55(2):153-5. doi: 10.1016/j.molimm.2012.10.013. Epub 2012 Nov 2. Mol Immunol. 2013. PMID: 23123036 Review. - IRAP identifies an endosomal compartment required for MHC class I cross-presentation.
Saveanu L, Carroll O, Weimershaus M, Guermonprez P, Firat E, Lindo V, Greer F, Davoust J, Kratzer R, Keller SR, Niedermann G, van Endert P. Saveanu L, et al. Science. 2009 Jul 10;325(5937):213-7. doi: 10.1126/science.1172845. Epub 2009 Jun 4. Science. 2009. PMID: 19498108 - Conventional dendritic cells require IRAP-Rab14 endosomes for efficient cross-presentation.
Weimershaus M, Maschalidi S, Sepulveda F, Manoury B, van Endert P, Saveanu L. Weimershaus M, et al. J Immunol. 2012 Feb 15;188(4):1840-6. doi: 10.4049/jimmunol.1101504. Epub 2012 Jan 11. J Immunol. 2012. PMID: 22238454 - IRAP Endosomes Control Phagosomal Maturation in Dendritic Cells.
Weimershaus M, Mauvais FX, Evnouchidou I, Lawand M, Saveanu L, van Endert P. Weimershaus M, et al. Front Cell Dev Biol. 2020 Dec 11;8:585713. doi: 10.3389/fcell.2020.585713. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33425891 Free PMC article. - Beyond the proteasome: trimming, degradation and generation of MHC class I ligands by auxiliary proteases.
Saveanu L, Fruci D, van Endert P. Saveanu L, et al. Mol Immunol. 2002 Oct;39(3-4):203-15. doi: 10.1016/s0161-5890(02)00102-5. Mol Immunol. 2002. PMID: 12200051 Review.
Cited by
- Pathways of MHC I cross-presentation of exogenous antigens.
Cruz FM, Chan A, Rock KL. Cruz FM, et al. Semin Immunol. 2023 Mar;66:101729. doi: 10.1016/j.smim.2023.101729. Epub 2023 Feb 16. Semin Immunol. 2023. PMID: 36804685 Free PMC article. Review. - Targeted Analysis of Serum Proteins Encoded at Known Inflammatory Bowel Disease Risk Loci.
Drobin K, Assadi G, Hong MG, Andersson E, Fredolini C, Forsström B, Reznichenko A, Akhter T, Ek WE, Bonfiglio F, Hansen MB, Sandberg K, Greco D, Repsilber D, Schwenk JM, D'Amato M, Halfvarson J. Drobin K, et al. Inflamm Bowel Dis. 2019 Jan 10;25(2):306-316. doi: 10.1093/ibd/izy326. Inflamm Bowel Dis. 2019. PMID: 30358838 Free PMC article. - Molecular and cell-biological mechanisms of antigen cross-presentation.
Kurts C. Kurts C. Front Immunol. 2013 Feb 28;4:51. doi: 10.3389/fimmu.2013.00051. eCollection 2013. Front Immunol. 2013. PMID: 23450983 Free PMC article. No abstract available. - IRAP Inhibitors: M1-Aminopeptidase Family Inspiration.
Barlow N, Thompson PE. Barlow N, et al. Front Pharmacol. 2020 Sep 25;11:585930. doi: 10.3389/fphar.2020.585930. eCollection 2020. Front Pharmacol. 2020. PMID: 33101040 Free PMC article. Review. - Association of Polymorphisms of Metabolism-Related Genes with Psoriasis Vulgaris in Han Chinese.
Liu Y, Li X, Huang YP, Cui ZY, Bao J, Zhang XL, Guo Y, Su MJ, Lv XX, Han JW. Liu Y, et al. Biomed Res Int. 2021 Sep 18;2021:9920631. doi: 10.1155/2021/9920631. eCollection 2021. Biomed Res Int. 2021. PMID: 34589554 Free PMC article.
References
- Albiston A. L., Fernando R. N., Yeatman H. R., Burns P., Ng L., Daswani D., Diwakarla S., Pham V., Chai S. Y. (2010). Gene knockout of insulin-regulated aminopeptidase: loss of the specific binding site for angiotensin IV and age-related deficit in spatial memory. Neurobiol. Learn. Mem. 93, 19–3010.1016/j.nlm.2009.07.011 - DOI - PubMed
- Albiston A. L., Mcdowall S. G., Matsacos D., Sim P., Clune E., Mustafa T., Lee J., Mendelsohn F. A., Simpson R. J., Connolly L. M., Chai S. Y. (2001). Evidence that the angiotensin IV (AT(4)) receptor is the enzyme insulin-regulated aminopeptidase. J. Biol. Chem. 276, 48623–4862610.1074/jbc.C100512200 - DOI - PubMed
- Albiston A. L., Morton C. J., Ng H. L., Pham V., Yeatman H. R., Ye S., Fernando R. N., De Bundel D., Ascher D. B., Mendelsohn F. A., Parker M. W., Chai S. Y. (2008). Identification and characterization of a new cognitive enhancer based on inhibition of insulin-regulated aminopeptidase. FASEB J. 22, 4209–421710.1096/fj.08-112227 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous