Chd2 interacts with H3.3 to determine myogenic cell fate - PubMed (original) (raw)
. 2012 Jun 29;31(13):2994-3007.
doi: 10.1038/emboj.2012.136. Epub 2012 May 8.
Seiji Okada, Daijiro Konno, Jun Odawara, Tomohiko Yoshimi, Saori Yoshimura, Hiromi Kumamaru, Hirokazu Saiwai, Toshiaki Tsubota, Hitoshi Kurumizaka, Koichi Akashi, Taro Tachibana, Anthony N Imbalzano, Yasuyuki Ohkawa
Affiliations
- PMID: 22569126
- PMCID: PMC3395093
- DOI: 10.1038/emboj.2012.136
Chd2 interacts with H3.3 to determine myogenic cell fate
Akihito Harada et al. EMBO J. 2012.
Abstract
Cell differentiation is mediated by lineage-determining transcription factors. We show that chromodomain helicase DNA-binding domain 2 (Chd2), a SNF2 chromatin remodelling enzyme family member, interacts with MyoD and myogenic gene regulatory sequences to specifically mark these loci via deposition of the histone variant H3.3 prior to cell differentiation. Directed and genome-wide analysis of endogenous H3.3 incorporation demonstrates that knockdown of Chd2 prevents H3.3 deposition at differentiation-dependent, but not housekeeping, genes and inhibits myogenic gene activation. The data indicate that MyoD determines cell fate and facilitates differentiation-dependent gene expression through Chd2-dependent deposition of H3.3 at myogenic loci prior to differentiation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
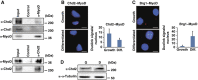
Chd2 interacts with MyoD. (A) Reciprocal IPs were performed from C2C12 myoblast extracts using MyoD- and Chd2-specific antibodies or IgG as a control. (B) PLAs indicating interaction of MyoD and Chd2 in both proliferating myoblasts and differentiated cells, in contrast to (C) the differentiation-specific interactions of MyoD and Brg1. Quantification represents the mean of three independent experiments, each of which analysed at least three separate fields±s.d. Scale bars=12.5 μm. (D) Western blot analysis of Chd2 levels in C2C12 cells under growth (G) or differentiation (D) conditions.
Figure 2
Chd2 interacts with MyoD and myogenic gene regulatory sequences. (A) ChIP assays for Chd2 binding at differentiation-dependent and skeletal muscle-specific (Acta1, Myl3, Myog, Cdkn1a, Ank1, Dmd), housekeeping (Gapdh, Ef1alpha), and silent (IgH enhancer, Pdx1, Neurod6) gene promoters were performed in C2C12 cells under growth and differentiated conditions. Relative recruitment was defined as the ratio of amplification of the PCR product relative to 1% of input genomic DNA. Values obtained from Acta1 at the growth stage were defined as 1 and all other values were expressed relative to that value. Each value was standardized by the amplification efficiency of each primer pair. Quantification represents the mean of three independent experiments±s.d. (B) Ectopic expression of MyoD induces Chd2 recruitment onto the promoter regions of myogenic genes. ChIP assays were performed as in (A) in fibroblast cells expressing MyoD or empty vector that were subjected to the differentiation protocol. (C) Western blot analysis for MyoD and Chd2 expression in MyoD-infected fibroblasts. H3 levels were monitored as a control. (D) siRNA-mediated MyoD knockdown inhibits Chd2 recruitment onto the promoter regions of myogenic genes. ChIP assays for Chd2 binding were performed as described in (A) in C2C12 myoblasts treated with either control siRNA or MyoD siRNA. (E) siRNA-mediated knockdown of the endogenous MyoD protein in C2C12 cells. A western blot analysis of C2C12 cells treated with either control siRNA or MyoD siRNA using antibodies against MyoD, Chd2, and H3 is shown. (F) Relative expression of skeletal muscle marker genes was reduced in C2C12 cells treated with MyoD siRNA. The levels of the indicated mRNAs were analysed by Q-PCR. The values in the differentiated cells expressing control siRNA were set to 1. Data represent the average of three independent experiments±s.d. (G) Chd2 and MyoD co-recruitment at the Ckm but not the Gapdh promoter is shown by re-ChIP. Re-ChIP experiments sequentially used antibodies against Chd2 and MyoD, as indicated. Relative recruitment was defined as the ratio of amplification of the PCR product relative to 1% of input genomic DNA. Values obtained from Ckm at the growth stage with 1st IP were defined as 1 and all other values were expressed relative to that value. Each value was standardized by the amplification efficiency of each primer pair. Quantification represents the mean of three independent experiments±s.d.
Figure 3
Chd2 is required for skeletal muscle differentiation. (A) The expression of myogenin (Myog) and myosin heavy chain (MHC) was not induced in C2C12 cells when Chd2 expression was suppressed by miRNAs targeting Chd2. To indirectly monitor miRNA expression, EGFP–NLS (EGFP fused with a nuclear transport signal) was expressed co-cistronically with miRNA. Scale bars=5 μm. (B) The transcription of skeletal muscle marker genes was suppressed in C2C12 cells expressing miRNA targeting Chd2. mRNA levels were analysed by Q-PCR; data represent the average of three independent experiments±s.d. Gapdh levels are shown as a control. (C) Western blot evaluating Chd2 protein knockdown and the expression of MyoD and other indicated proteins.
Figure 4
Myogenic phenotype is rescued by a forced expression of Chd2 partial mRNA in Chd2 knockdown cells. (A) The expression of myosin heavy chain (MHC; blue) was re-induced in C2C12 cells expressing miRNAs targeting Chd2 and a Chd2 partial mRNA that competes each miRNA. To indirectly monitor miRNA and Chd2 partial mRNA expression, EGFP–NLS (EGFP fused with a nuclear transport signal) and mKO1–NLS (mKO1 fused with a nuclear transport signal) were expressed co-cistronically with the miRNA and Chd2 partial mRNA, respectively. Scale bars=5 μm. Q-PCR analysis confirms equivalent expression of GFP and mKO1 in each sample (lower panel). (B) Chd2 expression was rescued in C2C12 cells expressing miRNAs targeting Chd2 and the competing Chd2 partial mRNA. Western blot analysis utilized antibodies against Chd2 and α-tubulin. (C) The transcription of skeletal muscle marker genes was rescued in C2C12 cells expressing miRNAs targeting Chd2 and the competing Chd2 partial mRNA. The levels of the indicated mRNAs were analysed by Q-PCR; data represent the average of three independent experiments±s.d.
Figure 5
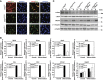
Chd2 interacts with H3.3 prior to and during skeletal muscle differentiation. (A) H3.3 antibody specifically discriminates between H3.3 and H3.1. Serial dilutions of purified recombinant H3.1 and H3.3 protein were evaluated by immunoblotting using the H3.3 and H3.1 monoclonal antibodies and the H3 polyclonal antibody. Detection of H3.3, H3.1, and H3 was performed on the same membrane. (B) IP from C2C12 cells using control IgG or Chd2-specific antibodies shows an interaction between Chd2 and H3.3. (C) PLAs indicating the frequency of interaction between H3.3 and Chd2 and between H3.1 and Chd2 in both proliferating myoblasts and differentiated cells. Quantification represents the mean of three independent experiments, each of which analysed at least three separate fields ±s.d. Scale bars=12.5 μm. (D) C2C12 cells at growth and differentiated stages were immunostained with antibodies to Chd2 (green) and H3.3 (red) to show Chd2 and H3.3 co-localization. Confocal images, cross-correlation analysis, and the amount of H3.3 interacting with Chd2 are shown. Scale bars=1 μm. (E) Co-immunoprecipitation from Chd2miR3139 cells expressing full-length Chd2–Flag or Chd2 deletion mutant (ΔChromodomain (281–512 aa)–Flag) using control IgG or Flag-antibodies shows an interaction between full-length Chd2 and H3.3.
Figure 6
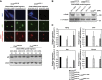
H3.3 incorporation at myogenic promoters occurs prior to differentiation and myogenic gene expression and is dependent on Chd2. (A) H3.3 incorporation at differentiation-specific myogenic gene promoters occurred prior to C2C12 cell differentiation. ChIP assays for myogenic, housekeeping, and silent genes were performed as described in Figure 2A. (B) Incorporation of H3.3 into myogenic gene loci is Chd2-dependent while incorporation housekeeping genes is Chd2-independent. ChIP assays for myogenic, housekeeping, and silent genes were in Chd2WT and Chd2miR (Chd2miR3139) expressing C2C12 cells under growth conditions as described in Figure 2A. (C) Chd2WT, Chd2miR3139, and Chd2miR5111-expressing C2C12 cells were cultured in growth medium for 1 day (G) and then shifted to differentiation medium for 48 h (D). H3.3 and total H3 expression levels were analysed by immunoblotting. (D) Ectopic expression of MyoD induces H3.3 incorporation at the promoter regions of myogenic genes. ChIP assays were performed as in Figure 2 in fibroblast cells expressing MyoD or empty vector. (E) siRNA-mediated MyoD knockdown inhibits H3.3 recruitment onto the promoter regions of myogenic, but not housekeeping, genes. ChIP assays were performed as in Figure 2 in C2C12 cells treated with either control siRNA or MyoD siRNA.
Figure 7
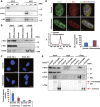
Knockdown of Chd2 decreases the incorporation of H3.3 into the muscle-specific gene loci on a genome-wide level. (A) H3.3 deposition in each chromosome is limited. The box plot represents the range of incorporation of H3.3 into each chromosome. (B) Chd2 knockdown changes the deposition of H3.3. The heat map represents the enrichment of H3.3 in upregulated genes during myogenic differentiation at the transcriptional start sites (TSS) ±5.5 kb. The maximum value of H3.3 enrichment is ∼10. Each row represents the enrichment pattern of H3.3 along the ±5.5 kb relative to the TSS. (C, D) Incorporation of H3.3 at the TSS and TES of muscle-specific, housekeeping, and silent gene loci in undifferentiated C2C12 cells. The H3.3 enrichment Tags from ChIP-Seq data aligned to the TSS and TES were segregated into 200 bp windows. Total enrichments were tallied in muscle-specific, housekeeping and silent genes (Supplementary Dataset 1). (E) MyoD-dependent induction of H3.3 incorporation at the TSS (left panel) and TES (right panel) of skeletal muscle gene loci in fibroblast cells ectopically expressing MyoD. Total enrichments were tallied for muscle-specific genes as in (C, D).
Figure 8
H3.3 is required for myogenic differentiation. (A) The expression of myosin heavy chain (MHC; red) and myotube formation was not induced in C2C12 cells when H3.3 expression was suppressed by siRNAs targeting H3f3a and H3f3b. Scale bars=5 μm. (B) The transcription of skeletal muscle marker genes was suppressed in C2C12 cells expressing siRNAs targeting H3f3a and H3f3b. mRNA levels were analysed by Q-PCR; data represent the average of three independent experiments±s.d. (C) Western blot evaluating H3.3 protein knockdown and the expression of the other indicated proteins. (D) siRNA-mediated H3.3 knockdown inhibits H3.3 recruitment onto Acta1, Myog and Myod1, while in Chd2 knockdown cells, H3.3 incorporation is not decreased at Myod1. ChIP assays were performed as in Figure 6. (E) Schematic representation of the ‘marking’ of myogenic genes by Chd2 prior to myogenesis. Chd2 coordinates with MyoD to direct H3.3 to differentiation-dependent, skeletal muscle-specific gene loci during the growth state. Incorporation of H3.3 marks myogenic loci for expression following the initiation of the differentiation process.
Similar articles
- Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition.
Yang JH, Song Y, Seol JH, Park JY, Yang YJ, Han JW, Youn HD, Cho EJ. Yang JH, et al. Proc Natl Acad Sci U S A. 2011 Jan 4;108(1):85-90. doi: 10.1073/pnas.1009830108. Epub 2010 Dec 20. Proc Natl Acad Sci U S A. 2011. PMID: 21173268 Free PMC article. - SH2B1 modulates chromatin state and MyoD occupancy to enhance expressions of myogenic genes.
Chen KW, Chang YJ, Yeh CM, Lian YL, Chan MW, Kao CF, Chen L. Chen KW, et al. Biochim Biophys Acta Gene Regul Mech. 2017 Feb;1860(2):270-281. doi: 10.1016/j.bbagrm.2016.12.007. Epub 2016 Dec 27. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 28039048 - The Scaffold attachment factor b1 (Safb1) regulates myogenic differentiation by facilitating the transition of myogenic gene chromatin from a repressed to an activated state.
Hernández-Hernández JM, Mallappa C, Nasipak BT, Oesterreich S, Imbalzano AN. Hernández-Hernández JM, et al. Nucleic Acids Res. 2013 Jun;41(11):5704-16. doi: 10.1093/nar/gkt285. Epub 2013 Apr 22. Nucleic Acids Res. 2013. PMID: 23609547 Free PMC article. - Epigenetic Regulation of Myogenesis: Focus on the Histone Variants.
Esteves de Lima J, Relaix F. Esteves de Lima J, et al. Int J Mol Sci. 2021 Nov 25;22(23):12727. doi: 10.3390/ijms222312727. Int J Mol Sci. 2021. PMID: 34884532 Free PMC article. Review. - Temporal regulation of chromatin during myoblast differentiation.
Harada A, Ohkawa Y, Imbalzano AN. Harada A, et al. Semin Cell Dev Biol. 2017 Dec;72:77-86. doi: 10.1016/j.semcdb.2017.10.022. Epub 2017 Oct 28. Semin Cell Dev Biol. 2017. PMID: 29079444 Free PMC article. Review.
Cited by
- Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture.
Sugimoto N, Maehara K, Yoshida K, Yasukouchi S, Osano S, Watanabe S, Aizawa M, Yugawa T, Kiyono T, Kurumizaka H, Ohkawa Y, Fujita M. Sugimoto N, et al. Nucleic Acids Res. 2015 Jul 13;43(12):5898-911. doi: 10.1093/nar/gkv509. Epub 2015 May 18. Nucleic Acids Res. 2015. PMID: 25990725 Free PMC article. - Sentinels of chromatin: chromodomain helicase DNA-binding proteins in development and disease.
Alendar A, Berns A. Alendar A, et al. Genes Dev. 2021 Nov 1;35(21-22):1403-1430. doi: 10.1101/gad.348897.121. Genes Dev. 2021. PMID: 34725129 Free PMC article. Review. - CHD1 and CHD2 are positive regulators of HIV-1 gene expression.
Rodgers MJ, Banks DJ, Bradley KA, Young JA. Rodgers MJ, et al. Virol J. 2014 Oct 8;11:180. doi: 10.1186/1743-422X-11-180. Virol J. 2014. PMID: 25297984 Free PMC article. - Interplay between PML NBs and HIRA for H3.3 dynamics following type I interferon stimulus.
Kleijwegt C, Bressac F, Seurre C, Bouchereau W, Cohen C, Texier P, Simonet T, Schaeffer L, Lomonte P, Corpet A. Kleijwegt C, et al. Elife. 2023 May 25;12:e80156. doi: 10.7554/eLife.80156. Elife. 2023. PMID: 37227756 Free PMC article. - Regulation of human cortical interneuron development by the chromatin remodeling protein CHD2.
Lewis EMA, Chapman G, Kaushik K, Determan J, Antony I, Meganathan K, Narasimhan M, Gontarz P, Zhang B, Kroll KL. Lewis EMA, et al. Sci Rep. 2022 Sep 17;12(1):15636. doi: 10.1038/s41598-022-19654-y. Sci Rep. 2022. PMID: 36115870 Free PMC article.
References
- Ahmad K, Henikoff S (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases