A resource for the conditional ablation of microRNAs in the mouse - PubMed (original) (raw)
. 2012 Apr 19;1(4):385-91.
doi: 10.1016/j.celrep.2012.02.008.
Lukas T Jeker, Karen Carver-Moore, Alyssia Oh, Huey Jiin Liu, Rachel Cameron, Hunter Richards, Zhongmei Li, David Adler, Yuko Yoshinaga, Maria Martinez, Michael Nefadov, Abul K Abbas, Art Weiss, Lewis L Lanier, Pieter J de Jong, Jeffrey A Bluestone, Deepak Srivastava, Michael T McManus
Affiliations
- PMID: 22570807
- PMCID: PMC3345170
- DOI: 10.1016/j.celrep.2012.02.008
A resource for the conditional ablation of microRNAs in the mouse
Chong Yon Park et al. Cell Rep. 2012.
Abstract
The importance of miRNAs during development and disease processes is well established. However, most studies have been done in cells or with patient tissues, and therefore the physiological roles of miRNAs are not well understood. To unravel in vivo functions of miRNAs, we have generated conditional, reporter-tagged knockout-first mice for numerous evolutionarily conserved miRNAs. Here, we report the generation of 162 miRNA targeting vectors, 64 targeted ES cell lines, and 46 germline-transmitted miRNA knockout mice. In vivo lacZ reporter analysis in 18 lines revealed highly tissue-specific expression patterns and their miRNA expression profiling matched closely with published expression data. Most miRNA knockout mice tested were viable, supporting a mechanism by which miRNAs act redundantly with other miRNAs or other pathways. These data and collection of resources will be of value for the in vivo dissection of miRNA functions in mouse models.
Conflict of interest statement
Present address: ES Cell Targeting Core, Research Resource Program, UCSF, San Francisco, CA 94158, USA
Figures
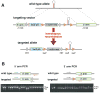
Figure 1. ES cell targeting and PCR genotyping
(A) Schematic representation of a miRNA locus and targeting strategy. Two homology arms at the 5′ and 3′ ends of the targeting vector mediate gene-specific targeting by homologous recombination. Targeting leads to insertions of a promoter-less lacZ reporter with an IRES (in black color) and a polyA signal, β-actin-driven neomycin selection marker with a polyA signal, and a miRNA stem-loop flanked by loxP sites into the miRNA locus. (B) PCR genotyping strategy of targeted ES cells. Positions of the PCR primers used for genotyping wild type and targeted alleles are marked with arrows and expected size differences for PCR products are illustrated. To verify homologous recombination on the 5′ arm, PCR was done with 2 gene-specific primers and 1 universal mutant primer. For the longer 3′arm side, PCR was done to verify the insertion of the third loxP site with 2 gene-specific primers. The mutant products were verified by sequencing.
Figure 2. Generation of knockout mice
(A) Breeding strategy. Combination of FRT and loxP sites allows manipulation of targeted allele in mice. Germline-transmitted mice (lacZ-neo-flox) can be crossed with germline deleter Flp mice, which will remove the reporter and the neomycin cassette to restore the wild type allele (conditional allele/flox). Then conditional mice can be crossed with either germline- or tissue-specific Cre transgenic mice to generate knockouts (knockout allele/KO). Instead of sequential recombination steps, germline-transmitted mice can be crossed with germline deleter Cre mice to produce mice with a reporter-tagged null allele (lacZ-knockout allele/lacZ-KO). (B) Flp-mediated recombination of FRT sites in mice. Germline-transmitted mice were crossed with homozygous Rosa-Flp mice. Parents and offspring were PCR-genotyped to check deletion of the lacZ reporter. Flp excision was not fully penetrant as the lacZ was not deleted in #5 pup, although it was completely excised in #2 pup (F and M indicate father and mother respectively). (C) Cre-mediated recombination of loxP sites in mice. Germline-transmitted mice were crossed with actin-Cre transgenic mice. Parents and offspring were PCR-genotyped to evaluate deletion of the neomycin marker, but not of the lacZ reporter. Excision by actin-Cre was complete among all pups (#7 to #9). (D) qPCR analysis to confirm the loss of miRNA in lacZ-KO mice. Either lung (miR-30b/30d, miR-339, miR-130a, miR-141/200c, miR-479/195 and miR-296/298) or brain (miR-7a-2) of 2 to 6 month-old knockout mice was used to isolate total RNA. RT-PCR was done by using Taqman miRNA assays. PCR were done as triplicates and data were presented as means with standard deviation.
Figure 3. Expression analysis of miRNA lacZ reporter
(A) LacZ reporter expression in E11.5 embryos. Embryos for miR-30b/30d, miR-30e, miR-130a, miR-296/298, miR-301a, and miR-339 displayed lacZ expression ubiquitously, whereas embryos for miR-210, miR-146a, mir-688, miR-497/195, and miR-654/376b exhibited no expression. The central nervous system was positive for miR-7a-2, miR-135b, and miR-325. Embryos for miR-141/200c showed staining only in the nostrils, as shown with arrows. Posterior trunk staining was detected for miR-196a-1. Although both expressed in pharyngeal arches and limb buds, expression patterns of miR-130b/301b and miR-205 were distinct. In addition, miR-130b/301b embryos displayed forebrain staining. (B) lacZ reporter expression in E18.5 embryos and adult. Brains of E18.5 embryos for miR-7a-2, miR-135b, and miR-325 displayed broad lacZ expression, whereas miR-688 exhibited very restricted expression domain in the dorsal cortex and ventral mid-brain (for each miRNA, top and bottom panels correspond to dorsal and ventral view respectively). Expression of miR-135b was also found in the inner ears (semicircular canals, vestibule and cochlea), while miR-325 expression was found in the visual sensory system (eyes, optic tracts, and lateral geniculate nucleus). The spinal cord staining was detected for miR-325. The CNS expression pattern of miR-7a-2 was maintained in adults as shown in the vibratome-sectioned brain. Expression of miR-141/200c was found in the airways of the respiratory tract, including in the nasal cavity, trachea, bronchi, and bronchioles, as well as in olfactory bulbs in the brain. Expression of miR-497/195 was found both in the airways and the air sacs of the lungs. Kidneys of miR-196a-1 displayed a striking lacZ staining pattern. For miR-654/376b, the frontal portion of the ribcage (corresponding to cartilage at this developmental stage) stained positive in E18.5 embryos.
Similar articles
- A resource of vectors and ES cells for targeted deletion of microRNAs in mice.
Prosser HM, Koike-Yusa H, Cooper JD, Law FC, Bradley A. Prosser HM, et al. Nat Biotechnol. 2011 Aug 7;29(9):840-5. doi: 10.1038/nbt.1929. Nat Biotechnol. 2011. PMID: 21822254 Free PMC article. - Integrated miRNA and mRNA expression profiling of mouse mammary tumor models identifies miRNA signatures associated with mammary tumor lineage.
Zhu M, Yi M, Kim CH, Deng C, Li Y, Medina D, Stephens RM, Green JE. Zhu M, et al. Genome Biol. 2011 Aug 16;12(8):R77. doi: 10.1186/gb-2011-12-8-r77. Genome Biol. 2011. PMID: 21846369 Free PMC article. - Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells.
Mallanna SK, Rizzino A. Mallanna SK, et al. Dev Biol. 2010 Aug 1;344(1):16-25. doi: 10.1016/j.ydbio.2010.05.014. Epub 2010 May 15. Dev Biol. 2010. PMID: 20478297 Free PMC article. Review. - Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?
Gunaratne PH. Gunaratne PH. Curr Stem Cell Res Ther. 2009 Sep;4(3):168-77. doi: 10.2174/157488809789057400. Curr Stem Cell Res Ther. 2009. PMID: 19492978 Review. - Transgene-Like Animal Models Using Intronic MicroRNAs.
Lin SL, Chang SE, Ying SY. Lin SL, et al. Methods Mol Biol. 2018;1733:239-254. doi: 10.1007/978-1-4939-7601-0_20. Methods Mol Biol. 2018. PMID: 29435938
Cited by
- The Pitx2:miR-200c/141:noggin pathway regulates Bmp signaling and ameloblast differentiation.
Cao H, Jheon A, Li X, Sun Z, Wang J, Florez S, Zhang Z, McManus MT, Klein OD, Amendt BA. Cao H, et al. Development. 2013 Aug;140(16):3348-59. doi: 10.1242/dev.089193. Epub 2013 Jul 17. Development. 2013. PMID: 23863486 Free PMC article. - Short-term memory of danger signals and environmental stimuli in immune cells.
Monticelli S, Natoli G. Monticelli S, et al. Nat Immunol. 2013 Aug;14(8):777-84. doi: 10.1038/ni.2636. Nat Immunol. 2013. PMID: 23867934 - MicroRNA-mediated regulation of T helper cell differentiation and plasticity.
Baumjohann D, Ansel KM. Baumjohann D, et al. Nat Rev Immunol. 2013 Sep;13(9):666-78. doi: 10.1038/nri3494. Epub 2013 Aug 2. Nat Rev Immunol. 2013. PMID: 23907446 Free PMC article. Review. - Arf tumor suppressor and miR-205 regulate cell adhesion and formation of extraembryonic endoderm from pluripotent stem cells.
Li C, Finkelstein D, Sherr CJ. Li C, et al. Proc Natl Acad Sci U S A. 2013 Mar 19;110(12):E1112-21. doi: 10.1073/pnas.1302184110. Epub 2013 Mar 4. Proc Natl Acad Sci U S A. 2013. PMID: 23487795 Free PMC article. - A transgenic resource for conditional competitive inhibition of conserved Drosophila microRNAs.
Fulga TA, McNeill EM, Binari R, Yelick J, Blanche A, Booker M, Steinkraus BR, Schnall-Levin M, Zhao Y, DeLuca T, Bejarano F, Han Z, Lai EC, Wall DP, Perrimon N, Van Vactor D. Fulga TA, et al. Nat Commun. 2015 Jun 17;6:7279. doi: 10.1038/ncomms8279. Nat Commun. 2015. PMID: 26081261 Free PMC article.
References
- Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials