Transcriptional and post-transcriptional regulation of SPAST, the gene most frequently mutated in hereditary spastic paraplegia - PubMed (original) (raw)
Transcriptional and post-transcriptional regulation of SPAST, the gene most frequently mutated in hereditary spastic paraplegia
Brian J Henson et al. PLoS One. 2012.
Abstract
Hereditary spastic paraplegias (HSPs) comprise a group of neurodegenerative disorders that are characterized by progressive spasticity of the lower extremities, due to axonal degeneration in the corticospinal motor tracts. HSPs are genetically heterogeneous and show autosomal dominant inheritance in ∼70-80% of cases, with additional cases being recessive or X-linked. The most common type of HSP is SPG4 with mutations in the SPAST gene, encoding spastin, which occurs in 40% of dominantly inherited cases and in ∼10% of sporadic cases. Both loss-of-function and dominant-negative mutation mechanisms have been described for SPG4, suggesting that precise or stoichiometric levels of spastin are necessary for biological function. Therefore, we hypothesized that regulatory mechanisms controlling expression of SPAST are important determinants of spastin biology, and if altered, could contribute to the development and progression of the disease. To examine the transcriptional and post-transcriptional regulation of SPAST, we used molecular phylogenetic methods to identify conserved sequences for putative transcription factor binding sites and miRNA targeting motifs in the SPAST promoter and 3'-UTR, respectively. By a variety of molecular methods, we demonstrate that SPAST transcription is positively regulated by NRF1 and SOX11. Furthermore, we show that miR-96 and miR-182 negatively regulate SPAST by effects on mRNA stability and protein level. These transcriptional and miRNA regulatory mechanisms provide new functional targets for mutation screening and therapeutic targeting in HSP.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Transcriptional regulation of the SPAST gene encoding spastin (SPG4). A
) Cartoon showing the human SPAST promoter structure with _cis_-elements representing putative transcription factor (TF) binding sites for NRF1, SOX11, and Sp1. As is typical of CpG-promoters, transcription start sites (TSS) are spread over a large region in exon 1, with two major TSS positions indicated by arrows . There are two alternative translational initiation codons for spastin 68 and 60 kDa polypeptide isoforms, respectively . B) Multi-sequence alignment of conserved TF _cis_-elements in representative mammalian species. Sequences were aligned using ClustalW 2.1 and manually adjusted as needed for maximum parsimony. Evolutionarily conserved TF motifs are indicated; *, nucleotide positions conserved in all 19 species; ∧, nucleotide positions conserved in 17/19 species; yellow shading, highly conserved SOX11 motif; red, NRF1 motifs; purple, Sp1 motifs. The NRF1 and SOX11 motifs are highly conserved, but only one Sp1 motif near the 5′ TSS is conserved in mammals. Extended alignments of the complete promoter region into exon 1 and including the first translational start codon are shown in Fig. S1.
Figure 2. NRF1 chromatin immunoprecipitation (ChIP) assays.
NRF1 interacts with the SPAST promoter in A) human SK-N-SH cells, and B) murine Neuro2a cells, by ChIP assay. The _SNURF_-SNRPN enhancer for human and _Snurf_-Snrpn promoter for mouse are positive controls for _cis_-elements known to be bound by NRF1 . Chromatin was immunoprecipitated with anti-NRF1 antibodies, and the promoter regions were assessed by PCR. Controls are no antibody (no Ab) and total input DNA (TI) for ChIP, and a water control (H2O) for PCR.
Figure 3. siRNA targeting NRF1 mRNA knocks down SPAST
mRNA expression. SK-N-SH cells were transfected with a pSUPER-NRF1 shRNA expression vector or mock transfected, with qualitative analysis of gene expression including a negative control (GAPDH). The quantitative band intensities for the “siRNA” and “mock” transfection samples were compared and the ratio is listed to the right of the gel images. The GAPDH mRNA level is unaffected by siRNA targeting NRF1, whereas the mRNA levels for NRF1 and SPAST are reduced by the siRNA treatment. The experiment was repeated three times with equivalent results to the representative results shown here.
Figure 4. siRNA targeting NRF1 mRNA ablates SPAST promoter function.
A) Cartoon showing the structure of the pGL3e-SPAST-promoter-luciferase vector, and the inhibitory mechanisms of siRNA action. Symbols are as for Fig. 1A. B) The plasmid pGL3e-SPAST-promoter was co-transfected into SK-N-SH cells with pSUPER shRNA vectors that target either NRF1, luciferase, or negative control (Arl2). *, P<0.05.
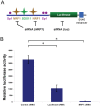
Figure 5. Regulation of the human SPAST promoter by SOX11.
A) Luciferase reporter assays with the human SPAST promoter. The plasmid pGL3b-SPAST-promoter-Luc (third row) and two deletion derivatives (depicted in the panel on the left) were transfected into Flp-in293 cells and normalized to cells transfected with the pGL3b vector (fourth row). *, P<0.05. B) Over-expression of SOX11 upregulates SPAST promoter-reporter constructs having a SOX11 binding site. Flp-In-293 cells were transfected with _SPAST_-promoter-luciferase reporter constructs both with (blue) and without (red) the SOX11 expression vector. *, P<0.05; **, P<0.001; ***, P<0.0001, and n.s., not statistically significant. Symbols in A and B are as for Fig. 1A.
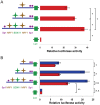
Figure 6. Over-expression of SOX11 and NRF1 upregulates endogenous SPAST expression.
Cells were transfected with SOX11 or NRF1 expression vectors and compared to cells transfected with the transfection control (pGL3b). By qRT-PCR, with normalization to GAPDH mRNA levels and to the transfection control, the levels of NRF1 and SOX11 mRNA were increased 182±28-fold and 2,925±241-fold on transfection with NRF1-VP16 and CMV1-SOX11, respectively (data not shown). Similarly, by qRT-PCR analysis, SPAST transcript levels are significantly increased by over-expression of SOX11 and NRF1. These data represent the average of three biological replicates each done in triplicate. *, P<0.05.
Figure 7. Prediction of several conserved seed motifs for miRNA targets in the SPAST 3′-UTR. A
) Cartoon of the SPAST mRNA showing positions of the coding sequence (green), polyA site (red), Alu repetitive element (gray) with target site duplication (black triangles), and miRNA target sites (purple) analyzed in this study. B) Base-pairing of Homo sapiens (hsa) miR-96 and miR-182 miRNAs with the SPAST 3′-UTR. The optimal seed motifs (red or brown) for targeting by the miRNAs are at position 511–517 (#1) and 462–467 (#2), respectively, of the SPAST 3′-UTR. C) Evolutionary conservation of the miR-96 target site in the SPAST 3′-UTR of mammals. The optimal target for the miR-96 seed (+2 to+8) is shown in blue, an optimal miR-182 seed in green, and other nucleotides that can base pair with miR-96 are in gray. Sequences conserved in all 38 species are indicated by *, and sequences conserved in 35 of 38 (90%) species by ∧. D) Evolutionary conservation of the miR-96 target site in the SPAST 3′-UTR of tetrapods. Footnotes: a Distance in nucleotides from stop codon to the first position in the target site for the miR-96 seed. b The optimal target for the miR-96 seed (+2 to+8) is shown in blue, other nucleotides that can base pair with miR-96 are in gray. c This distance corresponds to a manually corrected sequence, to overcome a poor sequence assembly in the database version.
Figure 8. Post-transcriptional regulation of SPAST.
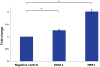
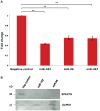
A) QRT-PCR data showing that miR-182, miR-96, and miR-367 reduce SPAST transcript levels in Flp-In-293 cells. These data represent the average of three biological replicates each done in triplicate. **, P<0.001. B) Western blot analysis of spastin and GAPDH protein expression in Flp-In-293 cells transfected with miR-96, miR-182, and the negative (neg) control (see Materials and Methods).
Similar articles
- Amplifying the spectrum of SPAST gene mutations.
Verriello L, Lonigro IR, Pessa ME, Betto E, Pauletto G, Fogolari F, Gigli GL, Curcio F. Verriello L, et al. Acta Biomed. 2021 Nov 18;92(S1):e2021220. doi: 10.23750/abm.v92iS1.11608. Acta Biomed. 2021. PMID: 35132972 Free PMC article. - A complex form of hereditary spastic paraplegia in three siblings due to somatic mosaicism for a novel SPAST mutation in the mother.
Aulitzky A, Friedrich K, Gläser D, Gastl R, Kubisch C, Ludolph AC, Volk AE. Aulitzky A, et al. J Neurol Sci. 2014 Dec 15;347(1-2):352-5. doi: 10.1016/j.jns.2014.09.046. Epub 2014 Oct 2. J Neurol Sci. 2014. PMID: 25315759 - Hereditary spastic paraplegia SPG4: what is known and not known about the disease.
Solowska JM, Baas PW. Solowska JM, et al. Brain. 2015 Sep;138(Pt 9):2471-84. doi: 10.1093/brain/awv178. Epub 2015 Jun 20. Brain. 2015. PMID: 26094131 Free PMC article. Review. - Genetic analysis of SPG4 and SPG3A genes in a cohort of Chinese patients with hereditary spastic paraplegia.
Lu X, Cen Z, Xie F, Ouyang Z, Zhang B, Zhao G, Luo W. Lu X, et al. J Neurol Sci. 2014 Dec 15;347(1-2):368-71. doi: 10.1016/j.jns.2014.10.017. Epub 2014 Oct 16. J Neurol Sci. 2014. PMID: 25454648 - A p.Arg499His mutation in SPAST is associated with infantile-onset complicated spastic paraplegia: a case report and review of the literature.
Nan H, Shiraku H, Mizuno T, Takiyama Y. Nan H, et al. BMC Neurol. 2021 Nov 9;21(1):439. doi: 10.1186/s12883-021-02478-0. BMC Neurol. 2021. PMID: 34753439 Free PMC article. Review.
Cited by
- Mutation in TECPR2 reveals a role for autophagy in hereditary spastic paraparesis.
Oz-Levi D, Ben-Zeev B, Ruzzo EK, Hitomi Y, Gelman A, Pelak K, Anikster Y, Reznik-Wolf H, Bar-Joseph I, Olender T, Alkelai A, Weiss M, Ben-Asher E, Ge D, Shianna KV, Elazar Z, Goldstein DB, Pras E, Lancet D. Oz-Levi D, et al. Am J Hum Genet. 2012 Dec 7;91(6):1065-72. doi: 10.1016/j.ajhg.2012.09.015. Epub 2012 Nov 21. Am J Hum Genet. 2012. PMID: 23176824 Free PMC article. - Elk1 affects katanin and spastin proteins via differential transcriptional and post-transcriptional regulations.
Kelle D, Kırımtay K, Selçuk E, Karabay A. Kelle D, et al. PLoS One. 2019 Feb 21;14(2):e0212518. doi: 10.1371/journal.pone.0212518. eCollection 2019. PLoS One. 2019. PMID: 30789974 Free PMC article. - The lncRNA MALAT1/miR-30/Spastin Axis Regulates Hippocampal Neurite Outgrowth.
Jiang T, Cai Z, Ji Z, Zou J, Liang Z, Zhang G, Liang Y, Lin H, Tan M. Jiang T, et al. Front Cell Neurosci. 2020 Oct 20;14:555747. doi: 10.3389/fncel.2020.555747. eCollection 2020. Front Cell Neurosci. 2020. PMID: 33192306 Free PMC article. - Gait Patterns in Patients with Hereditary Spastic Paraparesis.
Serrao M, Rinaldi M, Ranavolo A, Lacquaniti F, Martino G, Leonardi L, Conte C, Varrecchia T, Draicchio F, Coppola G, Casali C, Pierelli F. Serrao M, et al. PLoS One. 2016 Oct 12;11(10):e0164623. doi: 10.1371/journal.pone.0164623. eCollection 2016. PLoS One. 2016. PMID: 27732632 Free PMC article. - The Role of Spastin in Axon Biology.
Costa AC, Sousa MM. Costa AC, et al. Front Cell Dev Biol. 2022 Jul 5;10:934522. doi: 10.3389/fcell.2022.934522. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35865632 Free PMC article. Review.
References
- Depienne C, Stevanin G, Brice A, Durr A. Hereditary spastic paraplegias: an update. Curr Opin Neurol. 2007;20:674–680. - PubMed
- Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7:1127–1138. - PubMed
- Fink JK. In GeneReviews, ed. Pagon RA, Bird TC, Dolan CR, Stephens K, University of Washington, Seattle; 2009. Hereditary spastic paraplegia overview. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources