Release and utilization of N-acetyl-D-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp. infantis - PubMed (original) (raw)
Release and utilization of N-acetyl-D-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp. infantis
Daniel Garrido et al. Anaerobe. 2012 Aug.
Abstract
Human milk contains high amounts of complex oligosaccharides, which can be utilized especially by Bifidobacterium species in the infant gut as a carbon and energy source. N-acetyl-D-glucosamine is a building block of these oligosaccharides, and molecular details on the release and utilization of this monosaccharide are not fully understood. In this work we have studied some of the enzymatic properties of three N-acetyl-β-D-hexosaminidases encoded by the genome of the intestinal isolate Bifidobacterium longum subsp. infantis ATCC 15697 and the gene expression of the corresponding genes during bacterial growth on human milk oligosaccharides. These enzymes belong to the glycosyl hydrolase family 20, with several homologs in bifidobacteria. Their optimum pH was 5.0 and optimum temperature was 37 °C. The three enzymes were active on the GlcNAcβ1-3 linkage found in lacto-N-tetraose, the most abundant human milk oligosaccharide. Blon_0459 and Blon_0732, but not Blon_2355, cleaved branched GlcNAcβ1-6 linkages found in lacto-N-hexaose, another oligosaccharide abundant in breast milk. Bifidobacterium infantis N-acetyl-β-D-hexosaminidases were induced during early growth in vitro on human milk oligosaccharides, and also during growth on lacto-N-tetraose or lacto-N-neotetraose. The up-regulation of enzymes that convert this monosaccharide into UDP-N-acetylglucosamine by human milk oligosaccharides suggested that this activated sugar is used in peptidoglycan biosynthesis. These results emphasize the complexity of human milk oligosaccharide consumption by this infant intestinal isolate, and provide new clues into this process.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
Fig. 1.
Thin layer chromatography of co-incubations of B. infantis N-acetyl-β-D-hexosaminidases with LNT or LNH after treatment with β-galactosidases. Structures are illustrated below the figure. Lanes 1–8 and 26–28: standards (as indicated in the figure); lane 9: LNT with specific β1-3 galactosidase; lanes 10–12: LNT with a β1-3 galactosidase and Blon_0459, Blon_0732 or Blon_2355. Lane 13: LNH; lanes 14–16: LNH with either a β1-3, a β1-4 or both specific galactosidases; lanes 17–19: LNH with a β1-3 galactosidase and Blon_0459, Blon_0732 and Blon_2355, respectively; lanes 20–22: LNH with a β1-4 galactosidase and either Blon_0459, Blon_0732 and Blon_2355; lanes 23–25: LNH with both β1-3 and β1-4 galactosidase, as well as either Blon_0459, Blon_0732 and Blon_2355.
Fig. 2.
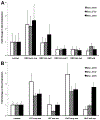
Relative quantification of the gene expression levels of β-hexosaminidase genes at different time points during growth on HMO (A), LNT or LNnT (B). Expression is relative to their levels during growth on lactose. Error bars represent two biological replicates.
Fig. 3.
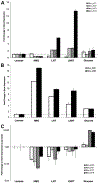
Fold change in gene expression for genes predicted to participate in the metabolism of GlcNAc in B. infantis after exponential growth on the substrates listed in the _x_-axis. Results are normalized to levels on lactose (dashed line), and error bars represent three biological replicates. A: Genes in the LNB/GNB pathway; B: GlcNAc-6-P deacetylase and GlcN-6-P isomerase; C: (results are presented in log scale) a PTS system putative for GlcNAc import and metabolism.
Similar articles
- Human Milk Oligosaccharide Utilization in Intestinal Bifidobacteria Is Governed by Global Transcriptional Regulator NagR.
Arzamasov AA, Nakajima A, Sakanaka M, Ojima MN, Katayama T, Rodionov DA, Osterman AL. Arzamasov AA, et al. mSystems. 2022 Oct 26;7(5):e0034322. doi: 10.1128/msystems.00343-22. Epub 2022 Sep 12. mSystems. 2022. PMID: 36094076 Free PMC article. - Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense.
Bunesova V, Lacroix C, Schwab C. Bunesova V, et al. BMC Microbiol. 2016 Oct 26;16(1):248. doi: 10.1186/s12866-016-0867-4. BMC Microbiol. 2016. PMID: 27782805 Free PMC article. - Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression.
Sakurama H, Kiyohara M, Wada J, Honda Y, Yamaguchi M, Fukiya S, Yokota A, Ashida H, Kumagai H, Kitaoka M, Yamamoto K, Katayama T. Sakurama H, et al. J Biol Chem. 2013 Aug 30;288(35):25194-25206. doi: 10.1074/jbc.M113.484733. Epub 2013 Jul 10. J Biol Chem. 2013. PMID: 23843461 Free PMC article. - Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides.
Kitaoka M. Kitaoka M. Adv Nutr. 2012 May 1;3(3):422S-9S. doi: 10.3945/an.111.001420. Adv Nutr. 2012. PMID: 22585921 Free PMC article. Review. - Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut.
Underwood MA, German JB, Lebrilla CB, Mills DA. Underwood MA, et al. Pediatr Res. 2015 Jan;77(1-2):229-35. doi: 10.1038/pr.2014.156. Epub 2014 Oct 10. Pediatr Res. 2015. PMID: 25303277 Free PMC article. Review.
Cited by
- Determining the metabolic fate of human milk oligosaccharides: it may just be more complex than you think?
Jackson PPJ, Wijeyesekera A, Rastall RA. Jackson PPJ, et al. Gut Microbiome (Camb). 2022 Sep 7;3:e9. doi: 10.1017/gmb.2022.8. eCollection 2022. Gut Microbiome (Camb). 2022. PMID: 39295778 Free PMC article. Review. - Integrative genomic reconstruction of carbohydrate utilization networks in bifidobacteria: global trends, local variability, and dietary adaptation.
Arzamasov AA, Rodionov DA, Hibberd MC, Guruge JL, Kazanov MD, Leyn SA, Kent JE, Sejane K, Bode L, Barratt MJ, Gordon JI, Osterman AL. Arzamasov AA, et al. bioRxiv [Preprint]. 2024 Jul 7:2024.07.06.602360. doi: 10.1101/2024.07.06.602360. bioRxiv. 2024. PMID: 39005317 Free PMC article. Preprint. - Variation in the Conservation of Species-Specific Gene Sets for HMO Degradation and Its Effects on HMO Utilization in Bifidobacteria.
Hermes GDA, Rasmussen C, Wellejus A. Hermes GDA, et al. Nutrients. 2024 Jun 15;16(12):1893. doi: 10.3390/nu16121893. Nutrients. 2024. PMID: 38931248 Free PMC article. - Genome-scale metabolic modeling of the human milk oligosaccharide utilization by Bifidobacterium longum subsp. infantis.
Román L, Melis-Arcos F, Pröschle T, Saa PA, Garrido D. Román L, et al. mSystems. 2024 Mar 19;9(3):e0071523. doi: 10.1128/msystems.00715-23. Epub 2024 Feb 16. mSystems. 2024. PMID: 38363147 Free PMC article. - Gastroprotective Effects of Oral Glycosaminoglycans with Sodium Alginate in an Indomethacin-Induced Gastric Injury Model in Rats.
Traserra S, Cuerda H, Vallejo A, Segarra S, Sabata R, Jimenez M. Traserra S, et al. Vet Sci. 2023 Nov 23;10(12):667. doi: 10.3390/vetsci10120667. Vet Sci. 2023. PMID: 38133218 Free PMC article.
References
- Le Huerou-Luron I, Blat S, Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev 2010;23:23–36. - PubMed
- Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr 2009;98:229–38. - PubMed
- Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr 2000;20:699–722. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HD061923/HD/NICHD NIH HHS/United States
- R01 HD065122/HD/NICHD NIH HHS/United States
- R01HD065122/HD/NICHD NIH HHS/United States
- R01HD061923/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources