Clinical application of exome sequencing in undiagnosed genetic conditions - PubMed (original) (raw)
Clinical application of exome sequencing in undiagnosed genetic conditions
Anna C Need et al. J Med Genet. 2012 Jun.
Abstract
Background: There is considerable interest in the use of next-generation sequencing to help diagnose unidentified genetic conditions, but it is difficult to predict the success rate in a clinical setting that includes patients with a broad range of phenotypic presentations.
Methods: The authors present a pilot programme of whole-exome sequencing on 12 patients with unexplained and apparent genetic conditions, along with their unaffected parents. Unlike many previous studies, the authors did not seek patients with similar phenotypes, but rather enrolled any undiagnosed proband with an apparent genetic condition when predetermined criteria were met.
Results: This undertaking resulted in a likely genetic diagnosis in 6 of the 12 probands, including the identification of apparently causal mutations in four genes known to cause Mendelian disease (TCF4, EFTUD2, SCN2A and SMAD4) and one gene related to known Mendelian disease genes (NGLY1). Of particular interest is that at the time of this study, EFTUD2 was not yet known as a Mendelian disease gene but was nominated as a likely cause based on the observation of de novo mutations in two unrelated probands. In a seventh case with multiple disparate clinical features, the authors were able to identify homozygous mutations in EFEMP1 as a likely cause for macular degeneration (though likely not for other features).
Conclusions: This study provides evidence that next-generation sequencing can have high success rates in a clinical setting, but also highlights key challenges. It further suggests that the presentation of known Mendelian conditions may be considerably broader than currently recognised.
Conflict of interest statement
Competing interests: None.
Figures
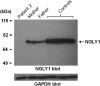
Figure 1
Expression of endogenous NGLY1 protein in peripheral blood mononuclear cells from patient, parents and three unrelated healthy controls. The protein expression level in the patient is less than both parents and healthy controls. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
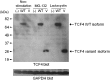
Figure 2
Expression of TCF4 variant and wild-type (WT) protein in COS-7 cells. The variant protein (V) is only seen in the presence of proteasome inhibitors. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
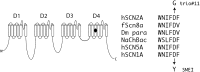
Figure 3
The SCN2A mutation, D1598G, is located in transmembrane segment 3 of the sodium channel protein domain 4. This residue is conserved in vertebrate, invertebrate DM (Drosophila) and bacterial (NaChBac) sodium channels. The D to Y mutation at the corresponding position of SCN1A was identified in a patient with severe myoclonic epilepsy (SME) of childhood, an early onset epileptic encephalopathy with features similar to the affected individual in trio 11 h, human; f, fish.
Similar articles
- What happens when N = 1 and you want plus 1?
Might M, Might CC. Might M, et al. Prenat Diagn. 2017 Jan;37(1):70-72. doi: 10.1002/pd.4975. Epub 2016 Dec 30. Prenat Diagn. 2017. PMID: 27885678 - Clinical exome sequencing for genetic identification of rare Mendelian disorders.
Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, Das K, Toy T, Harry B, Yourshaw M, Fox M, Fogel BL, Martinez-Agosto JA, Wong DA, Chang VY, Shieh PB, Palmer CG, Dipple KM, Grody WW, Vilain E, Nelson SF. Lee H, et al. JAMA. 2014 Nov 12;312(18):1880-7. doi: 10.1001/jama.2014.14604. JAMA. 2014. PMID: 25326637 Free PMC article. - Molecular findings among patients referred for clinical whole-exome sequencing.
Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, Veeraraghavan N, Hawes A, Chiang T, Leduc M, Beuten J, Zhang J, He W, Scull J, Willis A, Landsverk M, Craigen WJ, Bekheirnia MR, Stray-Pedersen A, Liu P, Wen S, Alcaraz W, Cui H, Walkiewicz M, Reid J, Bainbridge M, Patel A, Boerwinkle E, Beaudet AL, Lupski JR, Plon SE, Gibbs RA, Eng CM. Yang Y, et al. JAMA. 2014 Nov 12;312(18):1870-9. doi: 10.1001/jama.2014.14601. JAMA. 2014. PMID: 25326635 Free PMC article. - Exome sequencing identifies a de novo SCN2A mutation in a patient with intractable seizures, severe intellectual disability, optic atrophy, muscular hypotonia, and brain abnormalities.
Baasch AL, Hüning I, Gilissen C, Klepper J, Veltman JA, Gillessen-Kaesbach G, Hoischen A, Lohmann K. Baasch AL, et al. Epilepsia. 2014 Apr;55(4):e25-9. doi: 10.1111/epi.12554. Epub 2014 Mar 1. Epilepsia. 2014. PMID: 24579881 Review. - The Rise and Rise of Exome Sequencing.
Ku CS, Cooper DN, Patrinos GP. Ku CS, et al. Public Health Genomics. 2016;19(6):315-324. doi: 10.1159/000450991. Epub 2016 Nov 30. Public Health Genomics. 2016. PMID: 27898412 Review.
Cited by
- An overview of recent technological developments in bovine genomics.
Ghavi Hossein-Zadeh N. Ghavi Hossein-Zadeh N. Vet Anim Sci. 2024 Jul 23;25:100382. doi: 10.1016/j.vas.2024.100382. eCollection 2024 Sep. Vet Anim Sci. 2024. PMID: 39166173 Free PMC article. Review. - Identification and functional analysis of GJA8 mutation in a Chinese family with autosomal dominant perinuclear cataracts.
Su D, Yang Z, Li Q, Guan L, Zhang H, E D, Zhang L, Zhu S, Ma X. Su D, et al. PLoS One. 2013;8(3):e59926. doi: 10.1371/journal.pone.0059926. Epub 2013 Mar 29. PLoS One. 2013. PMID: 23555834 Free PMC article. - A case report of de novo missense FOXP1 mutation in a non-Caucasian patient with global developmental delay and severe speech impairment.
Song H, Makino Y, Noguchi E, Arinami T. Song H, et al. Clin Case Rep. 2015 Feb;3(2):110-3. doi: 10.1002/ccr3.167. Epub 2014 Nov 24. Clin Case Rep. 2015. PMID: 25767709 Free PMC article. - Development of new NGLY1 assay systems - toward developing an early screening method for NGLY1 deficiency.
Hirayama H, Fujihira H, Suzuki T. Hirayama H, et al. Glycobiology. 2024 Sep 30;34(11):cwae067. doi: 10.1093/glycob/cwae067. Glycobiology. 2024. PMID: 39206713 Free PMC article. Review. - Whole genome sequencing in support of wellness and health maintenance.
Patel CJ, Sivadas A, Tabassum R, Preeprem T, Zhao J, Arafat D, Chen R, Morgan AA, Martin GS, Brigham KL, Butte AJ, Gibson G. Patel CJ, et al. Genome Med. 2013 Jun 27;5(6):58. doi: 10.1186/gm462. eCollection 2013. Genome Med. 2013. PMID: 23806097 Free PMC article.
References
- Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, Basehore MJ, Broeckel U, Tomita-Mitchell A, Arca MJ, Casper JT, Margolis DA, Bick DP, Hessner MJ, Routes JM, Verbsky JW, Jacob HJ, Dimmock DP. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med 2011;13:255–62 - PubMed
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, van Bon BW, Hoischen A, de Vries BB, Brunner HG, Veltman JA. A de novo paradigm for mental retardation. Nat Genet 2010;42:1109–12 - PubMed
- Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, Dionne-Laporte A, Spiegelman D, Henrion E, Diallo O, Thibodeau P, Bachand I, Bao JY, Tong AH, Lin CH, Millet B, Jaafari N, Joober R, Dion PA, Lok S, Krebs MO, Rouleau GA. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet 2011;43:860–3 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 AG028377/AG/NIA NIH HHS/United States
- RC2 NS070342/NS/NINDS NIH HHS/United States
- R01 NS034509/NS/NINDS NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- NS34509/NS/NINDS NIH HHS/United States
- 1RC2NS070342-01/NS/NINDS NIH HHS/United States
- RC2 MH089915/MH/NIMH NIH HHS/United States
- RC2MH089915/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous