Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born - PubMed (original) (raw)
. 2012 May 8;8(5):1542-1555.
doi: 10.1021/ct200909j. Epub 2012 Mar 26.
- PMID: 22582031
- PMCID: PMC3348677
- DOI: 10.1021/ct200909j
Free PMC article
Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born
Andreas W Götz et al. J Chem Theory Comput. 2012.
Free PMC article
Abstract
We present an implementation of generalized Born implicit solvent all-atom classical molecular dynamics (MD) within the AMBER program package that runs entirely on CUDA enabled NVIDIA graphics processing units (GPUs). We discuss the algorithms that are used to exploit the processing power of the GPUs and show the performance that can be achieved in comparison to simulations on conventional CPU clusters. The implementation supports three different precision models in which the contributions to the forces are calculated in single precision floating point arithmetic but accumulated in double precision (SPDP), or everything is computed in single precision (SPSP) or double precision (DPDP). In addition to performance, we have focused on understanding the implications of the different precision models on the outcome of implicit solvent MD simulations. We show results for a range of tests including the accuracy of single point force evaluations and energy conservation as well as structural properties pertainining to protein dynamics. The numerical noise due to rounding errors within the SPSP precision model is sufficiently large to lead to an accumulation of errors which can result in unphysical trajectories for long time scale simulations. We recommend the use of the mixed-precision SPDP model since the numerical results obtained are comparable with those of the full double precision DPDP model and the reference double precision CPU implementation but at significantly reduced computational cost. Our implementation provides performance for GB simulations on a single desktop that is on par with, and in some cases exceeds, that of traditional supercomputers.
Figures
Figure 1
Peak floating-point operations per second (Flop/s; left) and memory bandwidth (right) for Intel CPUs and NVIDIA GPUs.
Figure 2
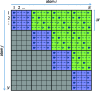
Schematic representation of the work-load distribution for the calculation of nonbond forces with N atoms. Each square represents the interactions between two atoms i and j for which the resulting forces need to be evaluated. These are grouped together in tiles of size W × W that are each assigned to an independent warp. Due to symmetry, only the blue diagonal tiles and the green off-diagonal tiles need to be considered for the calculation. For details, see the text.
Figure 3
Total energy (kcal/mol) along constant energy trajectories using a time step of 0.5 fs without constraints. Shown are results for TRPCage (left) and ubiquitin (right) for different precision models. The insets show the first nanosecond of each trajectory.
Figure 4
Root-mean-square deviations (RMSDs) of the Cα backbone carbon atoms of ubiquitin (excluding the flexible tail, residues 71–76) with respect to the crystal structure for 50 independent trajectories as obtained with the CPU implementation and the GPU implementation of PMEMD using different precision models.
Figure 5
Root-mean-square fluctuations (RMSFs) of the Cα backbone carbon atoms of ubiquitin residues 71–76 with respect to the crystal structure for 50 independent trajectories of 100 ns length as obtained with the CPU implementation and the GPU implementation of PMEMD using different precision models.
Similar articles
- Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald.
Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. Salomon-Ferrer R, et al. J Chem Theory Comput. 2013 Sep 10;9(9):3878-88. doi: 10.1021/ct400314y. Epub 2013 Aug 20. J Chem Theory Comput. 2013. PMID: 26592383 - GPU/CPU Algorithm for Generalized Born/Solvent-Accessible Surface Area Implicit Solvent Calculations.
Tanner DE, Phillips JC, Schulten K. Tanner DE, et al. J Chem Theory Comput. 2012 Jul 10;8(7):2521-2530. doi: 10.1021/ct3003089. Epub 2012 Jun 15. J Chem Theory Comput. 2012. PMID: 23049488 Free PMC article. - Fast and flexible gpu accelerated binding free energy calculations within the amber molecular dynamics package.
Mermelstein DJ, Lin C, Nelson G, Kretsch R, McCammon JA, Walker RC. Mermelstein DJ, et al. J Comput Chem. 2018 Jul 15;39(19):1354-1358. doi: 10.1002/jcc.25187. Epub 2018 Mar 12. J Comput Chem. 2018. PMID: 29532496 - Mixed-precision iterative refinement using tensor cores on GPUs to accelerate solution of linear systems.
Haidar A, Bayraktar H, Tomov S, Dongarra J, Higham NJ. Haidar A, et al. Proc Math Phys Eng Sci. 2020 Nov;476(2243):20200110. doi: 10.1098/rspa.2020.0110. Epub 2020 Nov 25. Proc Math Phys Eng Sci. 2020. PMID: 33363437 Free PMC article. - Stochastic rounding: implementation, error analysis and applications.
Croci M, Fasi M, Higham NJ, Mary T, Mikaitis M. Croci M, et al. R Soc Open Sci. 2022 Mar 9;9(3):211631. doi: 10.1098/rsos.211631. eCollection 2022 Mar. R Soc Open Sci. 2022. PMID: 35291325 Free PMC article. Review.
Cited by
- Multiple redox switches of the SARS-CoV-2 main protease in vitro provide opportunities for drug design.
Funk LM, Poschmann G, Rabe von Pappenheim F, Chari A, Stegmann KM, Dickmanns A, Wensien M, Eulig N, Paknia E, Heyne G, Penka E, Pearson AR, Berndt C, Fritz T, Bazzi S, Uranga J, Mata RA, Dobbelstein M, Hilgenfeld R, Curth U, Tittmann K. Funk LM, et al. Nat Commun. 2024 Jan 9;15(1):411. doi: 10.1038/s41467-023-44621-0. Nat Commun. 2024. PMID: 38195625 Free PMC article. - Fluorescence Polarization Screening Assays for Small Molecule Allosteric Modulators of ABL Kinase Function.
Grover P, Shi H, Baumgartner M, Camacho CJ, Smithgall TE. Grover P, et al. PLoS One. 2015 Jul 29;10(7):e0133590. doi: 10.1371/journal.pone.0133590. eCollection 2015. PLoS One. 2015. PMID: 26222440 Free PMC article. - Modeling the assembly of the multiple domains of α-actinin-4 and its role in actin cross-linking.
Travers T, Shao H, Wells A, Camacho CJ. Travers T, et al. Biophys J. 2013 Feb 5;104(3):705-15. doi: 10.1016/j.bpj.2012.12.003. Biophys J. 2013. PMID: 23442921 Free PMC article. - Speed of conformational change: comparing explicit and implicit solvent molecular dynamics simulations.
Anandakrishnan R, Drozdetski A, Walker RC, Onufriev AV. Anandakrishnan R, et al. Biophys J. 2015 Mar 10;108(5):1153-64. doi: 10.1016/j.bpj.2014.12.047. Biophys J. 2015. PMID: 25762327 Free PMC article. - Cyclic Peptide Inhibitors of the Tsg101 UEV Protein Interactions Refined through Global Docking and Gaussian Accelerated Molecular Dynamics Simulations.
Lin WW, Wang YJ, Ko CW, Cheng TL, Wang YT. Lin WW, et al. Polymers (Basel). 2020 Sep 28;12(10):2235. doi: 10.3390/polym12102235. Polymers (Basel). 2020. PMID: 32998394 Free PMC article.
References
- McCammon J. A.; Gelin B. R.; Karplus M. Nature 1977, 267, 585–590. - PubMed
- Duan Y.; Kollmann P. A. Science 1998, 282, 740–744. - PubMed
- Yeh I.-C.; Hummer G. J. Am. Chem. Soc. 2002, 124, 6563–6568. - PubMed
- Klepeis J. L.; Lindorff-Larsen K.; Dror R. O.; Shaw D. E. Curr. Opin. Struct. Biol. 2009, 19, 120–127. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources