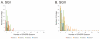Comparisons of clustered regularly interspaced short palindromic repeats and viromes in human saliva reveal bacterial adaptations to salivary viruses - PubMed (original) (raw)
Comparisons of clustered regularly interspaced short palindromic repeats and viromes in human saliva reveal bacterial adaptations to salivary viruses
David T Pride et al. Environ Microbiol. 2012 Sep.
Abstract
Explorations of human microbiota have provided substantial insight into microbial community composition; however, little is known about interactions between various microbial components in human ecosystems. In response to the powerful impact of viral predation, bacteria have acquired potent defences, including an adaptive immune response based on the clustered regularly interspaced short palindromic repeats (CRISPRs)/Cas system. To improve our understanding of the interactions between bacteria and their viruses in humans, we analysed 13 977 streptococcal CRISPR sequences and compared them with 2 588 172 virome reads in the saliva of four human subjects over 17 months. We found a diverse array of viruses and CRISPR spacers, many of which were specific to each subject and time point. There were numerous viral sequences matching CRISPR spacers; these matches were highly specific for salivary viruses. We determined that spacers and viruses coexist at the same time, which suggests that streptococcal CRISPR/Cas systems are under constant pressure from salivary viruses. CRISPRs in some subjects were just as likely to match viral sequences from other subjects as they were to match viruses from the same subject. Because interactions between bacteria and viruses help to determine the structure of bacterial communities, CRISPR-virus analyses are likely to provide insight into the forces shaping the human microbiome.
© 2012 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures
Figure 1
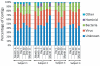
Putative biological assignments for contigs from human salivary viromes. Contigs were assigned to biological groups based on significant blastX E-scores based on the NCBI non-redundant database. The percentage of contigs assigned to each biological group is demonstrated for each subject at all time points. Subjects #1 and #2 were sampled at additional the additional time points of Month -6 and Month -3, however, no intervention took place during the study.
Figure 2
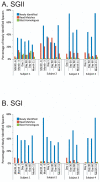
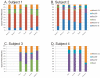
Percentage of spacers (Panels A and B) and diagram of SGII CRISPR spacer homologous to various bacteria, phage, and plasmids (Panel C). The percentage of spacers in each subject and time point that were not identified at prior time points is demonstrated in blue. The percentage of those spacers that match virome reads is demonstrated in red, and the percentage of those spacers that have homologues in the NCBI NR database are shown in green. Panel A - SGII spacers and Panel B - SGI spacers. For the initial time point for each subject, homologues to CRISPR spacer complements are demonstrated in Panel C. At each subsequent time point, only homologues to newly identified spacers that were not present in prior time points are shown. For example, in subject #3 (Panel C3), spacers homologous to streptococcal phage PH-10 are identified on Day 1, while on Day 30 newly identified spacers that were not present on Day 1 also have homology to phage PH-10. Panel C1-Subject #1, Panel C2 - Subject #2, Panel C3 - Subject #3, and Panel C4 - Subject #4. Homologues to phage are shown in red, plasmids in blue, and bacteria in green.
Figure 2
Percentage of spacers (Panels A and B) and diagram of SGII CRISPR spacer homologous to various bacteria, phage, and plasmids (Panel C). The percentage of spacers in each subject and time point that were not identified at prior time points is demonstrated in blue. The percentage of those spacers that match virome reads is demonstrated in red, and the percentage of those spacers that have homologues in the NCBI NR database are shown in green. Panel A - SGII spacers and Panel B - SGI spacers. For the initial time point for each subject, homologues to CRISPR spacer complements are demonstrated in Panel C. At each subsequent time point, only homologues to newly identified spacers that were not present in prior time points are shown. For example, in subject #3 (Panel C3), spacers homologous to streptococcal phage PH-10 are identified on Day 1, while on Day 30 newly identified spacers that were not present on Day 1 also have homology to phage PH-10. Panel C1-Subject #1, Panel C2 - Subject #2, Panel C3 - Subject #3, and Panel C4 - Subject #4. Homologues to phage are shown in red, plasmids in blue, and bacteria in green.
Figure 3
Fraction of virome reads with SGI CRISPR spacer matches over time within each subject. The virome reads with CRISPR spacer matches are normalized by virome size. Each column represents the SGI CRISPR spacer repertoire characterized at the individual labeled time point, and the Y axis represents the normalized percentages of unique reads with matches to CRISPR spacers recovered from each of the time points. Blue represents spacer-read matches from Month -6, red represents Month -3, green represents Day 1, purple represents Day 30, cyan represents Day 60, and orange represents Month 11.
Figure 4
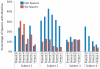
Percentage of spacers with matches to virome reads in each subject. The percentage of spacers that are unique to individual time points and those that are shared between multiple time points are shown. The percentage of spacers with matches to reads in each virome is demonstrated on the Y axis.
Figure 5
Heatmap of virome reads with CRISPR spacer matches for SGI and SGII CRISPR spacers. Each row represents a unique virome read, and each column represents the CRISPR spacer repertoire found on that day for that subject. Reads from viromes of each subject are identified on the left of each diagram, and the day from which each read was recovered is shown on the right of each diagram. The intensity scale bar is shown below each panel, and its values correspond to the percentage of CRISPR spacer matches present in each subject and time point. Panel A – SGI CRISPR spacer-read matches, Panel B – SGII CRISPR spacer-read matches.
Figure 6
Number of virome contigs with multiple SGI and SGII CRISPR spacer matches. The number of contigs is represented on the Y-axis, the number of CRISPR spacer matches for each contig is represented on the X-axis, and the different colors represent the subject from which each CRISPR spacer was derived.
Similar articles
- Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time.
Pride DT, Sun CL, Salzman J, Rao N, Loomer P, Armitage GC, Banfield JF, Relman DA. Pride DT, et al. Genome Res. 2011 Jan;21(1):126-36. doi: 10.1101/gr.111732.110. Epub 2010 Dec 13. Genome Res. 2011. PMID: 21149389 Free PMC article. - Diverse CRISPRs evolving in human microbiomes.
Rho M, Wu YW, Tang H, Doak TG, Ye Y. Rho M, et al. PLoS Genet. 2012;8(6):e1002441. doi: 10.1371/journal.pgen.1002441. Epub 2012 Jun 13. PLoS Genet. 2012. PMID: 22719260 Free PMC article. - Association between living environment and human oral viral ecology.
Robles-Sikisaka R, Ly M, Boehm T, Naidu M, Salzman J, Pride DT. Robles-Sikisaka R, et al. ISME J. 2013 Sep;7(9):1710-24. doi: 10.1038/ismej.2013.63. Epub 2013 Apr 18. ISME J. 2013. PMID: 23598790 Free PMC article. - Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes.
Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Al-Attar S, et al. Biol Chem. 2011 Apr;392(4):277-89. doi: 10.1515/BC.2011.042. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294681 Review. - Insights into the Human Virome Using CRISPR Spacers from Microbiomes.
Hidalgo-Cantabrana C, Sanozky-Dawes R, Barrangou R. Hidalgo-Cantabrana C, et al. Viruses. 2018 Sep 7;10(9):479. doi: 10.3390/v10090479. Viruses. 2018. PMID: 30205462 Free PMC article. Review.
Cited by
- Long-read metagenomics using PromethION uncovers oral bacteriophages and their interaction with host bacteria.
Yahara K, Suzuki M, Hirabayashi A, Suda W, Hattori M, Suzuki Y, Okazaki Y. Yahara K, et al. Nat Commun. 2021 Jan 4;12(1):27. doi: 10.1038/s41467-020-20199-9. Nat Commun. 2021. PMID: 33397904 Free PMC article. - Identification of Natural CRISPR Systems and Targets in the Human Microbiome.
Münch PC, Franzosa EA, Stecher B, McHardy AC, Huttenhower C. Münch PC, et al. Cell Host Microbe. 2021 Jan 13;29(1):94-106.e4. doi: 10.1016/j.chom.2020.10.010. Epub 2020 Nov 19. Cell Host Microbe. 2021. PMID: 33217332 Free PMC article. - Lysogeny in Streptococcus pneumoniae.
Garriss G, Henriques-Normark B. Garriss G, et al. Microorganisms. 2020 Oct 7;8(10):1546. doi: 10.3390/microorganisms8101546. Microorganisms. 2020. PMID: 33036379 Free PMC article. Review. - Filamentation initiated by Cas2 and its association with the acquisition process in cells.
Wang L, Yu X, Li M, Sun G, Zou L, Li T, Hou L, Guo Y, Shen D, Qu D, Cheng X, Chen L. Wang L, et al. Int J Oral Sci. 2019 Oct 3;11(3):29. doi: 10.1038/s41368-019-0063-0. Int J Oral Sci. 2019. PMID: 31578319 Free PMC article. - Oral microbiome and health.
Sharma N, Bhatia S, Sodhi AS, Batra N. Sharma N, et al. AIMS Microbiol. 2018 Jan 12;4(1):42-66. doi: 10.3934/microbiol.2018.1.42. eCollection 2018. AIMS Microbiol. 2018. PMID: 31294203 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources