Identification of a tetratricopeptide repeat-like domain in the nicastrin subunit of γ-secretase using synthetic antibodies - PubMed (original) (raw)
. 2012 May 29;109(22):8534-9.
doi: 10.1073/pnas.1202691109. Epub 2012 May 14.
Robert J Hoey, Guoqing Lin, Akiko Koide, Brenda Leung, Kwangwook Ahn, Georgia Dolios, Marcin Paduch, Takeshi Ikeuchi, Rong Wang, Yue-Ming Li, Shohei Koide, Sangram S Sisodia
Affiliations
- PMID: 22586122
- PMCID: PMC3365189
- DOI: 10.1073/pnas.1202691109
Identification of a tetratricopeptide repeat-like domain in the nicastrin subunit of γ-secretase using synthetic antibodies
Xulun Zhang et al. Proc Natl Acad Sci U S A. 2012.
Abstract
The γ-secretase complex, composed of presenilin, anterior-pharynx-defective 1, nicastrin, and presenilin enhancer 2, catalyzes the intramembranous processing of a wide variety of type I membrane proteins, including amyloid precursor protein (APP) and Notch. Earlier studies have revealed that nicastrin, a type I membrane-anchored glycoprotein, plays a role in γ-secretase assembly and trafficking and has been proposed to bind substrates. To gain more insights regarding nicastrin structure and function, we generated a conformation-specific synthetic antibody and used it as a molecular probe to map functional domains within nicastrin ectodomain. The antibody bound to a conformational epitope within a nicastrin segment encompassing residues 245-630 and inhibited the processing of APP and Notch substrates in in vitro γ-secretase activity assays, suggesting that a functional domain pertinent to γ-secretase activity resides within this region. Epitope mapping and database searches revealed the presence of a structured segment, located downstream of the previously identified DAP domain (DYIGS and peptidase; residues 261-502), that is homologous to a tetratricopeptide repeat (TPR) domain commonly involved in peptide recognition. Mutagenesis analyses within the predicted TPR-like domain showed that disruption of the signature helical structure resulted in the loss of γ-secretase activity but not the assembly of the γ-secretase and that Leu571 within the TPR-like domain plays an important role in mediating substrate binding. Taken together, these studies offer provocative insights pertaining to the structural basis for nicastrin function as a "substrate receptor" within the γ-secretase complex.
Conflict of interest statement
The authors declare no conflict of interest. S.S.S. is a paid consultant of Nociris Inc. and Eisai Research Labs Inc. but is not a shareholder in any company that is a maker or owner of a US Food and Drug Administration-regulated drug or device.
Figures
Fig. 1.
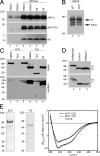
Analysis of deglycosylated NCT fragments. (A) HEK293 cells transiently transfected with either empty vector (lanes 1 and 2) or APPswe cDNA (lanes 3–6) were treated with either vehicle DMSO (lanes 1–4) or 1 μg/mL of kifunensine (kif). APP FL, CTF, and Aβ panels represent metabolites from either endogenous full-length APP (lanes 1 and 2) or overexpressed full-length APPswe (lanes 3–6). Duplicates are shown for each condition. (B) Analysis of Notch processing in naïve HEK293S cells (WT) or HEK293S GnTI− cells. Cells were transiently transfected with mouse NΔE cDNA, and full-length NΔE and NICD fragments are shown. (C) Analysis of ECD and 716 expressed in HEK293 cells. Western blots of lysates and medium of HEK293 cells stably expressing either ECD (lanes 1–3) or 716 (lanes 4–6) in the presence of vehicle DMSO, tunicamycin (Tuni), or kifunensine (kif). (D) Analysis of secreted ECD and 716 fragments from GnTI− cells in the presence or absence of Endo H (lane 2) or PNGaseF (lane 3). (E) Coomassie Blue staining of purified ECD and 716 fragments and circular dichroism spectroscopy of ECD and 716 from GnTI− cells or 716 from kifunensine-treated cell medium.
Fig. 2.
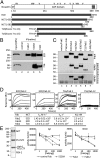
Potent NCT-specific Fab2 inhibits γ-secretase activity in vitro. (A) Schematic illustration of full-length human NCT and truncated NCT variants. SP, signal peptide; TM, transmembrane; TTR, transthyretin; 6HIS, His tag. (B) Immunoprecipitation of ECD (Upper) or 716 (Lower) from HEK293 cells with Fab12 (lanes 3), Mab5226A (_l_anes 4), or Fab2 (lanes 5). (C) Fab2 immunoprecipitation of secreted NCT fragments in medium of GnTI− cells (Top) in the absence (Middle) or presence (Bottom) of 0.1% Nonidet P-40. “con Fab” is a negative control Fab raised gainst an unrelated target. (D) SPR sensorgrams of Fab2 (ECD/Fab2 and 716/Fab2) or Fab12 (ECD/Fab12 and 716/Fab12) binding to immobilized ECD (Left and Center Left) or 716 (Right and Center Right); kinetic values are shown in the table. (E) Effects of Fabs on γ-secretase activity. Purified γ-secretase from HEK293S TAP-ANPP cells (Left). In vitro γ-secretase activity assays using purified γ-secretase preincubated with different Fabs; production of Aβ (Center) and NICD (Right) were monitored by ELISA. Data were represented as mean ± SEM, n = 3.
Fig. 3.
Sequence homology of a region of NCT ECD and TPR2A domain of HOP. (A) Western blot analysis of purified ECD and 716 probed with Fab12, Fab2, NCT55, and anti-6xHis antibodies. (B) Thermostability of ECD, Fab, and their complex was determined by differential scanning fluorimetry. (C) Alignment of NCT amino acid residues 501–669 to the TPR2A domain of HOP. The locations of helices of TPR2A domain are indicated above the alignment. (D) Homology modeling of TPR-like motif in ECD segment 501–669. Mutations in predicted α-helices in NCT 501–669 are labeled. The model is shown at two different angles to demonstrate the close proximity of L571 to the bound peptide (indicated as black line) in the model.
Fig. 4.
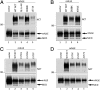
The TPR-like motif and L571 of NCT are important for γ-secretase activity. (A–D) NCT−/− cells were transiently cotransfected with mNΔE and cDNAs encoding NCT or various NCT mutants. Upper: Steady-state expression of NCT and NCT variants. Lower: Full-length mNΔE substrate and γ-secretase_–_generated NICD fragments. The identities of NCT variants are indicated.
Fig. 5.
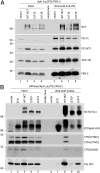
NCTs with the γ-secretase inactivating L571K or L571P mutations are recruited into the γ-secretase complex but fail to coimmunoprecipitate with APP CTFs. (A) NCT−/− cells were transiently cotransfected with Aph-1aL-Myc-His, PS1, PEN-2-CT11, and either WT or L571 mutant NCT cDNAs, and cell lysates were immunoprecipitated with anti-PS1 antibody. (B) NCT−/− cells were transiently cotransfected with APPswe, Aph-1aL-Myc-His, PS1, PEN2-CT11, and either WT, ECD, or L571 mutant NCT cDNAs. Anti-NCT Mab5226A was used to immunoprecipitate NCT and associated polypeptides. Immunoprecipitated NCT (NCT and ECD), APP CTFs (CTM1 and 26D6) and PS1 NTF are shown. Note that anti-His binding to full-length NCT in ECD panels is a residual signal from a reprobing of the CT11 immunoblot.
Similar articles
- Glu(332) in the Nicastrin ectodomain is essential for gamma-secretase complex maturation but not for its activity.
Chávez-Gutiérrez L, Tolia A, Maes E, Li T, Wong PC, de Strooper B. Chávez-Gutiérrez L, et al. J Biol Chem. 2008 Jul 18;283(29):20096-105. doi: 10.1074/jbc.M803040200. Epub 2008 May 23. J Biol Chem. 2008. PMID: 18502756 - A synthetic antibody fragment targeting nicastrin affects assembly and trafficking of γ-secretase.
Zhang X, Hoey R, Koide A, Dolios G, Paduch M, Nguyen P, Wu X, Li Y, Wagner SL, Wang R, Koide S, Sisodia SS. Zhang X, et al. J Biol Chem. 2014 Dec 12;289(50):34851-61. doi: 10.1074/jbc.M114.609636. Epub 2014 Oct 28. J Biol Chem. 2014. PMID: 25352592 Free PMC article. - Evidence That the "Lid" Domain of Nicastrin Is Not Essential for Regulating γ-Secretase Activity.
Zhang X, Sullivan E, Scimeca M, Wu X, Li YM, Sisodia SS. Zhang X, et al. J Biol Chem. 2016 Mar 25;291(13):6748-53. doi: 10.1074/jbc.C115.701649. Epub 2016 Feb 17. J Biol Chem. 2016. PMID: 26887941 Free PMC article. - Activity of gamma-secretase on substrates other than APP.
Lleó A. Lleó A. Curr Top Med Chem. 2008;8(1):9-16. doi: 10.2174/156802608783334060. Curr Top Med Chem. 2008. PMID: 18220928 Review. - The Alzheimer's disease-associated gamma-secretase complex: functional domains in the presenilin 1 protein.
Laudon H, Winblad B, Näslund J. Laudon H, et al. Physiol Behav. 2007 Sep 10;92(1-2):115-20. doi: 10.1016/j.physbeh.2007.05.037. Epub 2007 May 21. Physiol Behav. 2007. PMID: 17588625 Review.
Cited by
- Physiological and pathological roles of the γ-secretase complex.
Carroll CM, Li YM. Carroll CM, et al. Brain Res Bull. 2016 Sep;126(Pt 2):199-206. doi: 10.1016/j.brainresbull.2016.04.019. Epub 2016 Apr 28. Brain Res Bull. 2016. PMID: 27133790 Free PMC article. Review. - Curcumin Derivative GT863 Inhibits Amyloid-Beta Production via Inhibition of Protein N-Glycosylation.
Urano Y, Takahachi M, Higashiura R, Fujiwara H, Funamoto S, Imai S, Futai E, Okuda M, Sugimoto H, Noguchi N. Urano Y, et al. Cells. 2020 Feb 3;9(2):349. doi: 10.3390/cells9020349. Cells. 2020. PMID: 32028683 Free PMC article. - Substrate recruitment of γ-secretase and mechanism of clinical presenilin mutations revealed by photoaffinity mapping.
Fukumori A, Steiner H. Fukumori A, et al. EMBO J. 2016 Aug 1;35(15):1628-43. doi: 10.15252/embj.201694151. Epub 2016 May 23. EMBO J. 2016. PMID: 27220847 Free PMC article. - The dynamic conformational landscape of gamma-secretase.
Elad N, De Strooper B, Lismont S, Hagen W, Veugelen S, Arimon M, Horré K, Berezovska O, Sachse C, Chávez-Gutiérrez L. Elad N, et al. J Cell Sci. 2015 Feb 1;128(3):589-98. doi: 10.1242/jcs.164384. J Cell Sci. 2015. PMID: 25501811 Free PMC article. - Generating conformation-specific synthetic antibodies to trap proteins in selected functional states.
Paduch M, Koide A, Uysal S, Rizk SS, Koide S, Kossiakoff AA. Paduch M, et al. Methods. 2013 Mar 15;60(1):3-14. doi: 10.1016/j.ymeth.2012.12.010. Epub 2012 Dec 29. Methods. 2013. PMID: 23280336 Free PMC article.
References
- Price DL, Sisodia SS. Mutant genes in familial Alzheimer’s disease and transgenic models. Annu Rev Neurosci. 1998;21:479–505. - PubMed
- Tolia A, De Strooper B. Structure and function of gamma-secretase. Semin Cell Dev Biol. 2009;20:211–218. - PubMed
- Edbauer D, et al. Reconstitution of gamma-secretase activity. Nat Cell Biol. 2003;5:486–488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA062948/CA/NCI NIH HHS/United States
- P30 NS061777/NS/NINDS NIH HHS/United States
- R01 GM072688/GM/NIGMS NIH HHS/United States
- U54 GM087519/GM/NIGMS NIH HHS/United States
- S10 RR022415/RR/NCRR NIH HHS/United States
- U54-GM087519/GM/NIGMS NIH HHS/United States
- R01-GM072688/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases