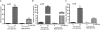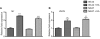Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7 - PubMed (original) (raw)
Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7
Melissa M Kendall et al. mBio. 2012.
Abstract
Bacterial pathogens must be able to both recognize suitable niches within the host for colonization and successfully compete with commensal flora for nutrients in order to establish infection. Ethanolamine (EA) is a major component of mammalian and bacterial membranes and is used by pathogens as a carbon and/or nitrogen source in the gastrointestinal tract. The deadly human pathogen enterohemorrhagic Escherichia coli O157:H7 (EHEC) uses EA in the intestine as a nitrogen source as a competitive advantage for colonization over the microbial flora. Here we show that EA is not only important for nitrogen metabolism but that it is also used as a signaling molecule in cell-to-cell signaling to activate virulence gene expression in EHEC. EA in concentrations that cannot promote growth as a nitrogen source can activate expression of EHEC's repertoire of virulence genes. The EutR transcription factor, known to be the receptor of EA, is only partially responsible for this regulation, suggesting that yet another EA receptor exists. This important link of EA with metabolism, cell-to-cell signaling, and pathogenesis, highlights the fact that a fundamental means of communication within microbial communities relies on energy production and processing of metabolites. Here we show for the first time that bacterial pathogens not only exploit EA as a metabolite but also coopt EA as a signaling molecule to recognize the gastrointestinal environment and promote virulence expression.
Importance: In order to successfully cause disease, a pathogen must be able to sense a host environment and modulate expression of its virulence genes as well as compete with the indigenous microbiota for nutrients. Ethanolamine (EA) is present in the large intestine due to the turnover of intestinal cells. Here, we show that the human pathogen Escherichia coli O157:H7, which causes bloody diarrhea and hemolytic-uremic syndrome, regulates virulence gene expression through EA metabolism and by responding to EA as a signal. These findings provide the first information directly linking EA with bacterial pathogenesis.
Figures
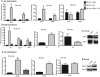
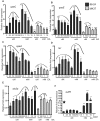
FIG 1
EA increases expression of EHEC virulence genes. (A) qRT-PCR of qseC and qseE of WT EHEC strain 86-24 grown in minimal medium containing glucose (glucose minimal medium) and either NH4, EA, or NH4 plus EA to early, mid-, and late log growth phases. (B) qRT-PCR of ler of WT EHEC grown in glucose minimal medium containing either NH4, EA, or NH4 plus EA to early, mid-, and late log phases. (C) Western blots of secreted proteins from WT EHEC grown to late log in glucose minimal medium containing either NH4 or EA probed with antisera against EspA from WT EHEC grown to late log. BSA, bovine serum albumin. (D) qRT-PCR of stx2a of WT EHEC grown in glucose minimal medium containing either NH4, EA, or NH4 plus EA to early, mid-, and late log. (E) Western blots of whole-cell lysates of WT EHEC grown in glucose minimal medium containing either NH4 or EA to mid log with antisera against Stx and RpoA (loading control). qRT-PCR expression values in panels A, B, and D are presented as relative values compared to the value for WT EHEC strain 86-24 grown with NH4. Values are means plus standard deviations (error bars) for 3 independent experiments. Values that are significantly different from the value for WT EHEC strain 86-24 grown with NH4 are indicated by asterisks as follows: *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
FIG 2
Expression of the eut operon genes eutR (A), eutB (B), and eutS (C) is induced in cells grown with EA in both low- and high-glucose media. qRT-PCR expression values are presented as relative values compared to WT EHEC strain 86-24 grown in low-glucose DMEM without EA. Error bars represent the standard deviations for 3 independent experiments. **, P < 0.005; ***, P < 0.0005.
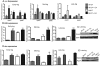
FIG 3
EA virulence gene regulation is both EutR dependent and independent. (A) qRT-PCR of qseC and qseE of WT EHEC strain 86-24 and the eutR mutant strain MK37 grown in DMEM or DMEM plus EA to early, mid-, and late log. (B) qRT-PCR of ler of WT EHEC strain 86-24 and the eutR mutant strain MK37 grown in DMEM or DMEM plus EA to early, mid-, and late log. (C) Western blots of secreted proteins from WT EHEC strain 86-24 and the eutR mutant strain MK37 grown to late log in DMEM or DMEM plus EA probed with antisera against EspA from WT EHEC. (D) qRT-PCR of stx2a of WT EHEC strain 86-24 and the eutR mutant strain MK37 grown in DMEM or DMEM plus EA to early, mid-, and late log. (E) Western blots of whole-cell lysates of WT EHEC strain 86-24 and the eutR mutant strain MK37 grown to late log in DMEM or DMEM plus EA with antisera against Stx and RpoA. RT-PCR expression values in panels A, B, and D are presented as relative values compared to WT EHEC strain 86-24 grown without EA. Error bars represent the standard deviations for 3 independent experiments. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
FIG 4
EA virulence gene regulation is independent of EA metabolism at early log growth phase. (A) qRT-PCR of ler of WT EHEC strain 86-24 and the eutB mutant strain MK47 grown in DMEM or DMEM plus EA. (B) qRT-PCR of stx2a of WT EHEC strain 86-24 and the eutB mutant strain MK47 grown in DMEM or DMEM plus EA. qRT-PCR expression values are presented as relative values compared to WT EHEC strain 86-24 grown without EA. Error bars represent the standard deviations for 3 independent experiments. **, P < 0.005; ***, P < 0.0005.
FIG 5
EA gene regulation in anaerobic conditions. (A) qRT-PCR of qseC in WT EHEC strain 86-24 and the eutR mutant strain MK37 in DMEM or DMEM plus EA. (B) qRT-PCR of qseE in WT EHEC strain 86-24 and the eutR mutant strain MK37 in DMEM or DMEM plus EA. (C) qRT-PCR of qseA in WT EHEC strain 86-24 and the eutR mutant strain MK37 in DMEM or DMEM plus EA. (D) qRT-PCR of ler in WT EHEC strain 86-24 and the eutR mutant strain MK37 in DMEM or DMEM plus EA. (E) qRT-PCR of stx2a in WT EHEC and eutR mutant strain MK37 in DMEM or DMEM plus EA. (F) qRT-PCR of eutR in WT EHEC grown in DMEM or DMEM plus EA and the eutR mutant strain MK37 grown in DMEM. The concentrations of EA (micromolar and millimolar) are shown on the x axes. qRT-PCR expression values are presented as relative values compared to WT EHEC strain 86-24 grown without EA. Error bars represent the standard deviations for 3 independent experiments. Statistical significance is indicated by symbols as follows. The asterisks indicate significance in gene expression between WT EHEC cells (0 µM EA), WT EHEC cells grown with EA, and the eutR mutant strain (0 µM EA): *, P < 0.05; **, P < 0.005; ***, P < 0.0005. The carets indicate significance in gene expression between the eutR mutant (0 µM EA) and eutR mutant cells grown with EA: ^, P < 0.05; ^^, P < 0.005; ^^^, P < 0.0005.
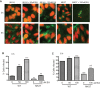
FIG 6
Detection of AE lesion formation using the FAS test on HeLa cells. (A) WT EHEC strain 86-24 and the eutR mutant strain MK37 in the presence and absence of EA at 3 h (top row) and 6 h (bottom row). The HeLa cell actin cytoskeleton (green) and the bacteria and HeLa cell nuclei (red) are shown. Pedestals are observed as bright green structures that are typically associated with bacterial cells. The cells were viewed at a magnification of ×640. The micromolar concentrations of EA are shown. (B and C) Percentage of infected HeLa cells after 3 h (B) or 6 h (C) of incubation. Error bars represent the standard deviations of 2 independent experiments. Statistical significance is indicated by symbols as follows. The asterisks indicate significance in gene expression between WT (0 µM EA), WT cells grown with EA, and the eutR mutant strain (0 µM EA): **, P < 0.005; ***, P < 0.0005. The carets indicate significance in gene expression between the eutR mutant cells (0 µM EA) and eutR mutant cells grown with EA; ^^, P < 0.005.
Comment in
- Ethanolamine: a signal to commence a host-associated lifestyle?
Garsin DA. Garsin DA. mBio. 2012 Jul 3;3(4):e00172-12. doi: 10.1128/mBio.00172-12. Print 2012. mBio. 2012. PMID: 22761393 Free PMC article.
Similar articles
- Ethanolamine Influences Human Commensal Escherichia coli Growth, Gene Expression, and Competition with Enterohemorrhagic E. coli O157:H7.
Rowley CA, Anderson CJ, Kendall MM. Rowley CA, et al. mBio. 2018 Oct 2;9(5):e01429-18. doi: 10.1128/mBio.01429-18. mBio. 2018. PMID: 30279284 Free PMC article. - EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7.
Luzader DH, Clark DE, Gonyar LA, Kendall MM. Luzader DH, et al. J Bacteriol. 2013 Nov;195(21):4947-53. doi: 10.1128/JB.00937-13. Epub 2013 Aug 30. J Bacteriol. 2013. PMID: 23995630 Free PMC article. - Ethanolamine: a signal to commence a host-associated lifestyle?
Garsin DA. Garsin DA. mBio. 2012 Jul 3;3(4):e00172-12. doi: 10.1128/mBio.00172-12. Print 2012. mBio. 2012. PMID: 22761393 Free PMC article. - Commensal 'trail of bread crumbs' provide pathogens with a map to the intestinal landscape.
Luzader DH, Kendall MM. Luzader DH, et al. Curr Opin Microbiol. 2016 Feb;29:68-73. doi: 10.1016/j.mib.2015.11.005. Epub 2015 Dec 17. Curr Opin Microbiol. 2016. PMID: 26707739 Free PMC article. Review. - SdiA sensing of acyl-homoserine lactones by enterohemorrhagic E. coli (EHEC) serotype O157:H7 in the bovine rumen.
Sperandio V. Sperandio V. Gut Microbes. 2010 Nov-Dec;1(6):432-5. doi: 10.4161/gmic.1.6.14177. Gut Microbes. 2010. PMID: 21468228 Free PMC article. Review.
Cited by
- The microbiota: a crucial mediator in gut homeostasis and colonization resistance.
Chen Y, Xiao L, Zhou M, Zhang H. Chen Y, et al. Front Microbiol. 2024 Aug 6;15:1417864. doi: 10.3389/fmicb.2024.1417864. eCollection 2024. Front Microbiol. 2024. PMID: 39165572 Free PMC article. Review. - Integrative in vivo analysis of the ethanolamine utilization bacterial microcompartment in Escherichia coli.
Jallet D, Soldan V, Shayan R, Stella A, Ismail N, Zenati R, Cahoreau E, Burlet-Schiltz O, Balor S, Millard P, Heux S. Jallet D, et al. mSystems. 2024 Aug 20;9(8):e0075024. doi: 10.1128/msystems.00750-24. Epub 2024 Jul 18. mSystems. 2024. PMID: 39023255 Free PMC article. - The yad and yeh fimbrial loci influence gene expression and virulence in enterohemorrhagic Escherichia coli O157:H7.
Gonyar LA, Sauder AB, Mortensen L, Willsey GG, Kendall MM. Gonyar LA, et al. mSphere. 2024 Jul 30;9(7):e0012424. doi: 10.1128/msphere.00124-24. Epub 2024 Jun 21. mSphere. 2024. PMID: 38904402 Free PMC article. - Role of ethanolamine utilization and bacterial microcompartment formation in Listeria monocytogenes intracellular infection.
Chatterjee A, Kaval KG, Garsin DA. Chatterjee A, et al. Infect Immun. 2024 Jun 11;92(6):e0016224. doi: 10.1128/iai.00162-24. Epub 2024 May 16. Infect Immun. 2024. PMID: 38752742 Free PMC article. - Ethanolamine enhances adhesion, promotes microcompartment formation, and modulates gene expression in Levilactobacillus brevis ATCC 14869.
Akouris PP, Stuivenberg GA, Chmiel JA, Kiattiburut W, Poon A, Reid G, Burton JP. Akouris PP, et al. Gut Microbes. 2024 Jan-Dec;16(1):2350778. doi: 10.1080/19490976.2024.2350778. Epub 2024 May 8. Gut Microbes. 2024. PMID: 38717446 Free PMC article.
References
- Bakovic M, Fullerton MD, Michel V. 2007. Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: the role of CTP:phosphoethanolamine cytidylyl-transferase (Pcyt2). Biochem. Cell Biol. 85:283–300 - PubMed
- Dowhan W. 1997. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 66:199–232 - PubMed
- Dowhan W. 1997. Phosphatidylserine decarboxylases: pyruvoyl-dependent enzymes from bacteria to mammals. Methods Enzymol. 280:81–88 - PubMed
- Snoeck V, Goddeeris B, Cox E. 2005. The role of enterocytes in the intestinal barrier function and antigen uptake. Microbes Infect. 7:997–1004 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical