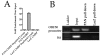KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome - PubMed (original) (raw)
KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome
Cyprian C Rossetto et al. PLoS Pathog. 2012.
Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is the cause of Kaposi's sarcoma and body cavity lymphomas. KSHV lytic infection produces PAN RNA, a highly abundant noncoding polyadenylated transcript that is retained in the nucleus. We recently demonstrated that PAN RNA interacts with several viral and cellular factors and can disregulate the expression of genes that modulate immune response. In an effort to define the role of PAN RNA in the context of the virus genome we generated a recombinant BACmid that deleted the PAN RNA locus. Because of the apparent duplication of the PAN RNA locus in BAC36, we generated BAC36CR, a recombinant BACmid that removes the duplicated region. BAC36CR was used as a template to delete most of the PAN RNA locus to generate BAC36CRΔPAN. BAC36CRΔPAN failed to produce supernatant virus and displayed a general decrease in mRNA accumulation of representative immediate early, early and late genes. Most strikingly, K-Rta expression was decreased in lytically induced BAC36CRΔPAN-containing cell lines at early and late time points post induction. Expression of PAN RNA in trans in BAC36CRΔPAN containing cells resulted in an increase in K-Rta expression, however K-Rta over expression failed to rescue BAC36CRΔPAN, suggesting that PAN RNA plays a wider role in virus replication. To investigate the role of PAN RNA in the activation of K-Rta expression, we demonstrate that PAN RNA physically interacts with the ORF50 promoter. RNA chromatin immunoprecipitation assays show that PAN RNA interacts with demethylases JMJD3 and UTX, and the histone methyltransferase MLL2. Consistent with the interaction with demethylases, expression of PAN RNA results in a decrease of the repressive H3K27me3 mark at the ORF50 promoter. These data support a model where PAN RNA is a multifunctional regulatory transcript that controls KSHV gene expression by mediating the modification of chromatin by targeting the KSHV repressed genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Deletion of the duplicated region within BAC36.
(A) Duplicated genomic region is located between two terminal repeat sequences of the BAC36 genome (B) The duplicated ORFs 18-K5 were removed by insertion of a GalK-KanR cassette using oligonucleotides homologous to regions outside of the duplicated region. (C) Replacement of the GalK-KanR cassette in the BAC36 genome to yield the recombinant BACmid BAC36CR where the entire duplicated regions was removed. Shown is the DNA sequence after removal of the cassette (D) Ethidium bromide stained gel and Southern blot of BAC36 and BAC36CR DNA cleaved with BamHI and hybridized with either a probe specific for the GalK-KanR cassette (GalK probe) or the PAN RNA locus (PAN probe). Lanes: 1, MW marker; 2, BAC36, 3, BAC36+GalK-KanR cassette; 4, BAC36CR. Arrows indicated the PAN RNA locus in the unique long region of the genome and the duplicated PAN RNA locus located between the terminal repeats.
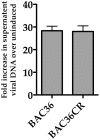
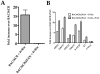
Figure 2. Production of infectious virus from BAC36CR induced cell lines.
BAC36 and BAC36CR cell lines were induced with TPA/n-butyrate for 5 days and supernatant virus was analyzed using qPCR. The amount of virus DNA accumulation was compared to DNA accumulation from uninduced BAC36. The error bars are the standard deviation of the mean from 3 separate experiments.
Figure 3. Generation of a recombinant BACmid with the PAN RNA locus deleted, BAC36CRΔPAN.
(A) The BAC36CR template was used to insert the GalK-KanR cassette such that 634 nts of the PAN RNA gene was removed from the genome. (B) The GalK-KanR cassette was removed by homologous recombination and reverse selection. (C) BAC36CRΔPAN was generated by removal of the GalK-KanR cassette and the putative polyadenlyation signal downstream of the original PAN RNA and K7 genes was preserved. (D) Ethidium bromide stained agarose gel and Southern blot of BAC36, BAC36CR and BAC36CRΔPAN DNA cleaved with BamHI showing the removal of part of the PAN RNA locus.
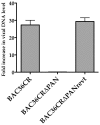
Figure 4. BAC36CRΔPAN fails to produce supernatant virus.
Supernatant virus was harvested from cell lines harboring BAC36CR, BAC36CRΔPAN or BAC36CRΔPANrevt DNA 5 days post induction. Relative amounts of viral DNA was analyzed using qPCR and is reported as fold increase in viral DNA level compared to uninduced BAC36CR.
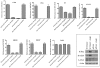
Figure 5. Decreased gene expression in the absence of PAN RNA expression.
qPCR analysis of mRNA accumulation from genes encoding K-Rta, K7, K-bZIP, ORF25 and ORF57 from BAC36CR, BAC36CRΔPAN or BAC36CRΔPANrevt cells treated with TPA/n-butyrate for 4 days. Values are compared to mRNA accumulation from uninduced BAC36CR containing cells. Each experiment was performed 3 times and error bars are the standard deviation from the mean. Boxed Panel. Western blot analysis of protein extracts reacted with anti-K-Rta, anti-K-bZIP, anti-LANA or anti-actin specific antibodies from induced and uninduced cell lines containing BAC36CR or BAC36CRΔPAN.
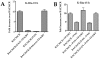
Figure 6. K-Rta mRNA accumulation is decreased at early times post induction in the absence of PAN RNA expression.
(A) qPCR analysis of K-Rta mRNA accumulation from BAC36CR or BAC36CRΔPAN containing cells treated with TPA/n-butyrate (TPA/BT) for 15 hr. The experiment was performed 3 times and error bars are the standard deviation from the mean. (B) Expression of PAN RNA in trans activates K-Rta transcription BACmid harboring cell lines. qPCR analysis of K-Rta mRNA accumulation from BAC36CR or BAC36CRΔPAN containing cells transfected with a PAN RNA expression plasmid with or without treatment with TPA/n-butyrate (TPA/BT). Total RNA was harvested 15 h post induction. The experiment was performed 3 times and error bars are the standard deviation from the mean.
Figure 7. Overexpression of K-Rta cannot complement BAC36CRΔPAN.
(A) BACmid containing cell lines were transfected with a K-Rta expression plasmid and supernatant virus DNA was measured 4 days post transfection. The experiment was repeated 3 times. Error bars are the standard deviation from the mean. (B) Trans expression of K-Rta activates viral promoters in BAC36CRΔPAN containing cells and expression is enhanced in the presence of PAN RNA. BAC36CRΔPAN containing cells were transfected with K-Rta with or without the cotransfection of the PAN RNA expression plasmid and qPCR analysis was performed to measure mRNA accumulation for several viral encoded genes.
Figure 8. PAN RNA physically interacts with the ORF50 promoter.
(A) PAN RNA is enriched 30-fold by ChIRP assay. PAN RNA or LacZ specific biotinylated oligonucleotides were used to enrich PAN RNA. Recovered RNA or RNA from the depleted lysate (post ChIRP) was measured by qPCR. (B) TREx/BCBL-1 Rta cells were treated with DOX and 3 days post treatment ChIRP assays were performed. Tiling biotinylated oligonucleotides were used that hybridized to either PAN RNA (20 oligonucleotides) or control LacZ RNA (20 oligonucleotides). Pulled down DNA that was occupied by RNA was amplified using primers specific for the ORF50 promoter region or K6 ORF coding sequence.
Figure 9. JMJD3 and UTX demethylases interact with KSHV DNA in the presence of PAN RNA expression.
Cell lines containing BAC36CR or BAC36CRΔPAN were transfected with a K-Rta expression plasmid and ChIP assays were performed 3 days post transfection. Immunoprecipitations were performed using antibodies specific for JMJD3, UTX, K-Rta or an isotype specific antibody control. PCR primers specific for the ORF50 promoter or ORF45 were used to amplify immunoprecipitated DNA. Panel BAC36CRΔPAN+PAN: cells were transfected with both a K-Rta and PAN RNA expression plasmid.
Figure 10. PAN RNA interacts with demethylases and the histone methyltransferase MLL2.
(A) TREx/BCBL-1 Rta cells were treated with DOX and RNA CLIP assays were performed 3 days post treatment. PAN RNA-protein complexes were immunoprecipitated using anti-JMJD3, anti-UTX, anti-MML2 or isotype control antibodies. PCR primers were used to amplify (after RT) PAN RNA, ORF45 RNA or U1 RNA. Also shown is PCR amplification without a reverse transcriptase reaction (PAN no RT). (B) PAN RNA expression leads to a relative decrease in the H3K27me3 mark on the ORF50 promoter. BAC36CR or BAC36CRΔPAN containing cells were transfected with either a K-Rta expression plasmid and/or a plasmid expressing PAN RNA. ChIP assays were performed using anti-H3K27me3 specific antibody. Immunoprecipitated DNA was analyzed by qPCR normalized to input DNA. Data is reported as fold decrease compared to BAC36CR untreated samples. Error bars are the standard deviation of the mean from three separate experiments.
Similar articles
- KSHV ORF59 and PAN RNA Recruit Histone Demethylases to the Viral Chromatin during Lytic Reactivation.
Hiura K, Strahan R, Uppal T, Prince B, Rossetto CC, Verma SC. Hiura K, et al. Viruses. 2020 Apr 9;12(4):420. doi: 10.3390/v12040420. Viruses. 2020. PMID: 32283586 Free PMC article. - PAN RNA: transcriptional exhaust from a viral engine.
Campbell M, Izumiya Y. Campbell M, et al. J Biomed Sci. 2020 Mar 7;27(1):41. doi: 10.1186/s12929-020-00637-y. J Biomed Sci. 2020. PMID: 32143650 Free PMC article. Review. - PAN's Labyrinth: Molecular biology of Kaposi's sarcoma-associated herpesvirus (KSHV) PAN RNA, a multifunctional long noncoding RNA.
Rossetto CC, Pari GS. Rossetto CC, et al. Viruses. 2014 Nov 4;6(11):4212-26. doi: 10.3390/v6114212. Viruses. 2014. PMID: 25375885 Free PMC article. Review.
Cited by
- A cultured affair: HSV latency and reactivation in neurons.
Wilson AC, Mohr I. Wilson AC, et al. Trends Microbiol. 2012 Dec;20(12):604-11. doi: 10.1016/j.tim.2012.08.005. Epub 2012 Sep 7. Trends Microbiol. 2012. PMID: 22963857 Free PMC article. Review. - Clinical Manifestations and Epigenetic Regulation of Oral Herpesvirus Infections.
Atyeo N, Rodriguez MD, Papp B, Toth Z. Atyeo N, et al. Viruses. 2021 Apr 15;13(4):681. doi: 10.3390/v13040681. Viruses. 2021. PMID: 33920978 Free PMC article. Review. - Gammaherpesvirus small noncoding RNAs are bifunctional elements that regulate infection and contribute to virulence in vivo.
Diebel KW, Oko LM, Medina EM, Niemeyer BF, Warren CJ, Claypool DJ, Tibbetts SA, Cool CD, Clambey ET, van Dyk LF. Diebel KW, et al. mBio. 2015 Feb 17;6(1):e01670-14. doi: 10.1128/mBio.01670-14. mBio. 2015. PMID: 25691585 Free PMC article. - Molecular biology of KSHV lytic reactivation.
Purushothaman P, Uppal T, Verma SC. Purushothaman P, et al. Viruses. 2015 Jan 14;7(1):116-53. doi: 10.3390/v7010116. Viruses. 2015. PMID: 25594835 Free PMC article. Review. - Kaposi's Sarcoma-Associated Herpesvirus mRNA Accumulation in Nuclear Foci Is Influenced by Viral DNA Replication and Viral Noncoding Polyadenylated Nuclear RNA.
Vallery TK, Withers JB, Andoh JA, Steitz JA. Vallery TK, et al. J Virol. 2018 Jun 13;92(13):e00220-18. doi: 10.1128/JVI.00220-18. Print 2018 Jul 1. J Virol. 2018. PMID: 29643239 Free PMC article.
References
- Al-Maghrabi JA. Castleman's disease. Update on pathogenesis. Saudi Med J. 2011;32:451–458. - PubMed
- Martin JN. Kaposi sarcoma-associated herpesvirus/human herpesvirus 8 and Kaposi sarcoma. Adv Dent Res. 2011;23:76–78. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous