Age of the association between Helicobacter pylori and man - PubMed (original) (raw)
doi: 10.1371/journal.ppat.1002693. Epub 2012 May 10.
Bodo Linz, Robert P Bond, Martin Nieuwoudt, Himla Soodyall, Carina M Schlebusch, Steffi Bernhöft, James Hale, Sebastian Suerbaum, Lawrence Mugisha, Schalk W van der Merwe, Mark Achtman
Affiliations
- PMID: 22589724
- PMCID: PMC3349757
- DOI: 10.1371/journal.ppat.1002693
Age of the association between Helicobacter pylori and man
Yoshan Moodley et al. PLoS Pathog. 2012.
Abstract
When modern humans left Africa ca. 60,000 years ago (60 kya), they were already infected with Helicobacter pylori, and these bacteria have subsequently diversified in parallel with their human hosts. But how long were humans infected by H. pylori prior to the out-of-Africa event? Did this co-evolution predate the emergence of modern humans, spanning the species divide? To answer these questions, we investigated the diversity of H. pylori in Africa, where both humans and H. pylori originated. Three distinct H. pylori populations are native to Africa: hpNEAfrica in Afro-Asiatic and Nilo-Saharan speakers, hpAfrica1 in Niger-Congo speakers and hpAfrica2 in South Africa. Rather than representing a sustained co-evolution over millions of years, we find that the coalescent for all H. pylori plus its closest relative H. acinonychis dates to 88-116 kya. At that time the phylogeny split into two primary super-lineages, one of which is associated with the former hunter-gatherers in southern Africa known as the San. H. acinonychis, which infects large felines, resulted from a later host jump from the San, 43-56 kya. These dating estimates, together with striking phylogenetic and quantitative human-bacterial similarities show that H. pylori is approximately as old as are anatomically modern humans. They also suggest that H. pylori may have been acquired via a single host jump from an unknown, non-human host. We also find evidence for a second Out of Africa migration in the last 52,000 years, because hpEurope is a hybrid population between hpAsia2 and hpNEAfrica, the latter of which arose in northeast Africa 36-52 kya, after the Out of Africa migrations around 60 kya.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
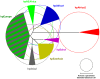
Figure 1. Neighbor-joining population tree of extant populations of H. pylori.
Circle diameters are proportional to the within-population genetic diversity (π). Angles of filled arcs are proportional to the number of isolates. Data are from , – and the figure is modified from Figure 1 in .
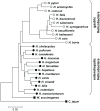
Figure 2. Neighbor-joining tree of 16SrRNA sequences from gastric and enterohepatic Helicobacter species.
Sequences from gastric Helicobacters (open circles) such as H. pylori, H. cetorum or H. suis form a cluster separate from enterohepatic Helicobacter species (filled circles) such as H. cinaedi or H. macacae. The tree was rooted with the 16SrRNA sequences of the epsilon-proteobacteria Wolinella succinogenes and Campylobacter jejuni (squares). 16SrRNA sequences were obtained from Genbank, accession numbers are provided in Materials and Methods.
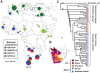
Figure 3. The distribution of H. pylori populations in Africa.
(A) The proportions of haplotypes at each sampling location (numbers; Table 2, Table S1) from different bacterial populations are displayed as pie charts whose sizes indicate the numbers of haplotypes. (B) The distribution of the three major subgroups of the San language family in south-central Africa (adapted from [21]). s: Southern Khoisan was spoken on much of the South African plateau and the central Kalahari in Botswana; c: Central Khoisan was distributed in southern and western South Africa, most of Namibia and most of northern Botswana; and n: Northern Khoisan (Ju), was spoken in southern Angola, north-eastern Namibia and north-western Botswana (Table 1). The position of the letters indicates the geographical origin of Northern San (n; !Xun from Angola), Central San (c; Khwe from Namibia) and Southern San (s; Khomani from South Africa). (C) Phylogenetic relationships among hpAfrica2 strains (80% consensus of 100 C
lonal
F
rame
analyses). The tree was rooted with H. pylori strains from other populations.
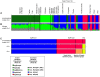
Figure 4. Bayesian population assignments using Structure V2.0.
(A) D
istruct
plot of the assignment of H. pylori haplotypes from Africa, the Middle East and southern Europe as determined by the no admixture model. Each isolate is represented by a thin line that is color coded according to the population assignment. (B) D
istruct
plot of the proportions of ancestral nucleotides as determined by the linkage model. A thin line for each isolate indicates the estimated amount of ancestry from each of the four ancestral populations as four colored segments. (C) D
istruct
plot of the population assignment (no admixture model) of H. pylori hpAfrica1 and hpAfrica2 haplotypes from southern Africa and H. acinonychis (Hac).
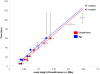
Figure 5. Linear relationships between known calibration dates and posterior parameters node height (ClonalFrame) and t (IMa).
Circles in red show C
lonal
F
rame
estimates and those in blue are IM
a
values. Closed circles denote median values for calibration times used for the regression (Table 3). Open circles show inferred times for the four African pair-wise comparisons (Table 4) after calibration with rate-smoothing.
Figure 6. A comparison of global H. pylori and human mtDNA phylogenies.
(A) Global phylogeny of H. pylori displayed as the strict consensus of 100 C
lonal
F
rame
analyses. After outgroup-rooting with H. cetorum, the time of the basal H. pylori divergence between hpAfrica2 and all other populations was estimated to 102 kya (95% confidence limit, 88–116 kya). Divergence of the other African H. pylori populations, hpAfrica1 and hpNEAfrica, began between 36 and 52 kya. (B) Simplified human mtDNA phylogeny adapted from Behar et al., 2008 with the permission of AJHG. African lineages are shown on a green background whereas the background for lineages outside Africa is light blue. San clades are purple, non-San clades are orange and H. acinonychis is yellow. San mtDNA lineages in our sample are shown as white lines.
Figure 7. Parallel patterns of pair-wise genetic distances between human mtDNA and H. pylori gene sequences.
A Mantel regression (R 2 = 0.62, P<0.0001) showed that 62% of the pair-wise genetic distances between H. pylori sequences can be accounted for by the pair-wise genetic distances between human mtDNA sequences from analogous geographic locations.
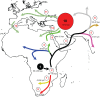
Figure 8. Chronological reconstruction of the major population events occurring during the intimate human-H. pylori association.
Black lines indicate undifferentiated populations and all other lines are color-coded according to population as in Figs. 1, 3A, 4A, 6A. The sequence of events is as follows: 1) Initial acquisition of H. pylori by a human ancestor; 2) Divergence of H. pylori into two super-lineages; 3) First successful migration of modern humans Out of Africa , via the southern route ; 4) H. pylori divergence into hpAfrica1 and hpNEAfrica with migration eastwards (hpNEAfrica) and westwards (hpAfrica1); 5) Divergence of H. pylori out of Africa into hpSahul and 6) hpAsia2 and hpEastAsia; 7) Host jump from San to large felines giving rise to H. acinonychis. 8) Southward migration of San carrying the ancestor of hpAfrica2; 9) Second successful migration Out of Africa via the Levant; 10) Hybridization of AE1 from central and south-west Asia and AE2 from north-east Africa in the Middle East or western Asia resulting in hpEurope; 11) Spread of hpEurope bacteria to Europe; 12) Back migration from the Middle East , and Spain spreading hpEurope into North Africa. Dates in italics represent estimates obtained from sources other than H. pylori.
Similar articles
- Recent acquisition of Helicobacter pylori by Baka pygmies.
Nell S, Eibach D, Montano V, Maady A, Nkwescheu A, Siri J, Elamin WF, Falush D, Linz B, Achtman M, Moodley Y, Suerbaum S. Nell S, et al. PLoS Genet. 2013;9(9):e1003775. doi: 10.1371/journal.pgen.1003775. Epub 2013 Sep 19. PLoS Genet. 2013. PMID: 24068950 Free PMC article. - An African origin for the intimate association between humans and Helicobacter pylori.
Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, Yamaoka Y, Graham DY, Perez-Trallero E, Wadstrom T, Suerbaum S, Achtman M. Linz B, et al. Nature. 2007 Feb 22;445(7130):915-918. doi: 10.1038/nature05562. Epub 2007 Feb 7. Nature. 2007. PMID: 17287725 Free PMC article. - Population genetic analyses of Helicobacter pylori isolates from Gambian adults and children.
Secka O, Moodley Y, Antonio M, Berg DE, Tapgun M, Walton R, Worwui A, Thomas V, Corrah T, Thomas JE, Adegbola RA. Secka O, et al. PLoS One. 2014 Oct 13;9(10):e109466. doi: 10.1371/journal.pone.0109466. eCollection 2014. PLoS One. 2014. PMID: 25310300 Free PMC article. - Relatedness of Helicobacter pylori populations to gastric carcinogenesis.
Dong QJ, Zhan SH, Wang LL, Xin YN, Jiang M, Xuan SY. Dong QJ, et al. World J Gastroenterol. 2012 Dec 7;18(45):6571-6. doi: 10.3748/wjg.v18.i45.6571. World J Gastroenterol. 2012. PMID: 23236231 Free PMC article. Review. - Helicobacter pylori infection in Africa: 2018 literature update.
Smith SI, Seriki A, Ndip R, Pellicano R. Smith SI, et al. Minerva Gastroenterol Dietol. 2018 Sep;64(3):222-234. doi: 10.23736/S1121-421X.18.02464-9. Epub 2018 Jan 10. Minerva Gastroenterol Dietol. 2018. PMID: 29327819 Review.
Cited by
- Allergies, Helicobacter pylori and the continental enigmas.
Sitaraman R. Sitaraman R. Front Microbiol. 2015 Jun 9;6:578. doi: 10.3389/fmicb.2015.00578. eCollection 2015. Front Microbiol. 2015. PMID: 26106380 Free PMC article. Review. - How old are bacterial pathogens?
Achtman M. Achtman M. Proc Biol Sci. 2016 Aug 17;283(1836):20160990. doi: 10.1098/rspb.2016.0990. Proc Biol Sci. 2016. PMID: 27534956 Free PMC article. Review. - Helicobacter pylori virulence factors affecting gastric proton pump expression and acid secretion.
Hammond CE, Beeson C, Suarez G, Peek RM Jr, Backert S, Smolka AJ. Hammond CE, et al. Am J Physiol Gastrointest Liver Physiol. 2015 Aug 1;309(3):G193-201. doi: 10.1152/ajpgi.00099.2015. Epub 2015 Jun 4. Am J Physiol Gastrointest Liver Physiol. 2015. PMID: 26045613 Free PMC article. - Pathogenomics of Helicobacter pylori.
Yamaoka Y, Saruuljavkhlan B, Alfaray RI, Linz B. Yamaoka Y, et al. Curr Top Microbiol Immunol. 2023;444:117-155. doi: 10.1007/978-3-031-47331-9_5. Curr Top Microbiol Immunol. 2023. PMID: 38231217 Review. - Worldwide Population Structure, Long-Term Demography, and Local Adaptation of Helicobacter pylori.
Montano V, Didelot X, Foll M, Linz B, Reinhardt R, Suerbaum S, Moodley Y, Jensen JD. Montano V, et al. Genetics. 2015 Jul;200(3):947-63. doi: 10.1534/genetics.115.176404. Epub 2015 May 20. Genetics. 2015. PMID: 25995212 Free PMC article.
References
- Suerbaum S, Michetti P. Helicobacter pylori infection. New England J Med. 2002;347:1175–1186. - PubMed
- Achtman M, Azuma T, Berg DE, Ito Y, Morelli G, et al. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical