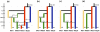New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties - PubMed (original) (raw)
doi: 10.1371/journal.pgen.1002703. Epub 2012 May 10.
Pierre Gladieux, Marinus J M Smulders, Isabel Roldán-Ruiz, François Laurens, Bruno Le Cam, Anush Nersesyan, Joanne Clavel, Marina Olonova, Laurence Feugey, Ivan Gabrielyan, Xiu-Guo Zhang, Maud I Tenaillon, Tatiana Giraud
Affiliations
- PMID: 22589740
- PMCID: PMC3349737
- DOI: 10.1371/journal.pgen.1002703
New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties
Amandine Cornille et al. PLoS Genet. 2012.
Abstract
The apple is the most common and culturally important fruit crop of temperate areas. The elucidation of its origin and domestication history is therefore of great interest. The wild Central Asian species Malus sieversii has previously been identified as the main contributor to the genome of the cultivated apple (Malus domestica), on the basis of morphological, molecular, and historical evidence. The possible contribution of other wild species present along the Silk Route running from Asia to Western Europe remains a matter of debate, particularly with respect to the contribution of the European wild apple. We used microsatellite markers and an unprecedented large sampling of five Malus species throughout Eurasia (839 accessions from China to Spain) to show that multiple species have contributed to the genetic makeup of domesticated apples. The wild European crabapple M. sylvestris, in particular, was a major secondary contributor. Bidirectional gene flow between the domesticated apple and the European crabapple resulted in the current M. domestica being genetically more closely related to this species than to its Central Asian progenitor, M. sieversii. We found no evidence of a domestication bottleneck or clonal population structure in apples, despite the use of vegetative propagation by grafting. We show that the evolution of domesticated apples occurred over a long time period and involved more than one wild species. Our results support the view that self-incompatibility, a long lifespan, and cultural practices such as selection from open-pollinated seeds have facilitated introgression from wild relatives and the maintenance of genetic variation during domestication. This combination of processes may account for the diversification of several long-lived perennial crops, yielding domestication patterns different from those observed for annual species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
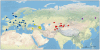
Figure 1. Geographic origins of the samples of the four wild Malus species used: M. sylvestris (blue), M. orientalis (yellow), M. baccata (purple), and M. sieversii (red).
Samples of unknown origin (N = 28) were not projected onto the map.
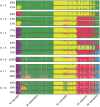
Figure 2. Proportions of ancestry of Malus genotypes from five species (N = 770) from K = 2 to K = 8 ancestral genepools (“clusters”) inferred with the STRUCTURE program.
Each individual is represented by a vertical bar, partitioned into K segments representing the amount of ancestry of its genome in K clusters. When several clustering solutions (“modes”) were represented within replicate runs, the proportion of simulations represented by each mode is given.
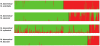
Figure 3. Proportions of ancestry in two ancestral genepools inferred with the STRUCTURE program, based on datasets including M. domestica (green, N = 299) and each of the four wild Malus species (red).
The x-axis is not to scale (details in Table S2).
Figure 4. Admixture models compared in approximate Bayesian computations.
Model a assumes that M. domestica is derived from M. sieversii and that the ancestral M. domestica population was involved in reciprocal introgression events with M. orientalis and M. sylvestris, and subsequently introgressed back into M. sieversii. Model b assumes no introgression from M. domestica into wild species, model c assumes the only admixture event is from M. sylvestris into M. domestica, and model d assumes no admixture. Admixture times between M. domestica and the three wild species were fixed (see text). Abbreviations: Nk, effective population sizes; Tk, divergence times; r1, r3, r4 introgression from M. domestica into M. sieversii, M. sylvestris, and M. orientalis respectively; r2, r5 introgression from M. sylvestris and M. orientalis, respectively, into M. domestica.
Figure 5. Proportions of ancestry of M. domestica genotypes (cider and dessert apples) in two ancestral genepools inferred with the STRUCTURE program.
Similar articles
- The domestication and evolutionary ecology of apples.
Cornille A, Giraud T, Smulders MJ, Roldán-Ruiz I, Gladieux P. Cornille A, et al. Trends Genet. 2014 Feb;30(2):57-65. doi: 10.1016/j.tig.2013.10.002. Epub 2013 Nov 27. Trends Genet. 2014. PMID: 24290193 Review. - The East Asian wild apples, Malus baccata (L.) Borkh and Malus hupehensis (Pamp.) Rehder., are additional contributors to the genomes of cultivated European and Chinese varieties.
Chen X, Cornille A, An N, Xing L, Ma J, Zhao C, Wang Y, Han M, Zhang D. Chen X, et al. Mol Ecol. 2023 Sep;32(18):5125-5139. doi: 10.1111/mec.16485. Epub 2022 May 24. Mol Ecol. 2023. PMID: 35510734 - Evolution of the population structure of Venturia inaequalis, the apple scab fungus, associated with the domestication of its host.
Gladieux P, Zhang XG, Róldan-Ruiz I, Caffier V, Leroy T, Devaux M, Van Glabeke S, Coart E, Le Cam B. Gladieux P, et al. Mol Ecol. 2010 Feb;19(4):658-74. doi: 10.1111/j.1365-294X.2009.04498.x. Epub 2010 Jan 18. Mol Ecol. 2010. PMID: 20088887 - Phenotypic divergence between the cultivated apple (Malus domestica) and its primary wild progenitor (Malus sieversii).
Davies T, Watts S, McClure K, Migicovsky Z, Myles S. Davies T, et al. PLoS One. 2022 Mar 23;17(3):e0250751. doi: 10.1371/journal.pone.0250751. eCollection 2022. PLoS One. 2022. PMID: 35320270 Free PMC article. - Genetic clues to the origin of the apple.
Harris SA, Robinson JP, Juniper BE. Harris SA, et al. Trends Genet. 2002 Aug;18(8):426-30. doi: 10.1016/s0168-9525(02)02689-6. Trends Genet. 2002. PMID: 12142012 Review.
Cited by
- Population Genomics of Domesticated Cucurbita ficifolia Reveals a Recent Bottleneck and Low Gene Flow with Wild Relatives.
Aguirre-Dugua X, Barrera-Redondo J, Gasca-Pineda J, Vázquez-Lobo A, López-Camacho A, Sánchez-de la Vega G, Castellanos-Morales G, Scheinvar E, Aguirre-Planter E, Lira-Saade R, Eguiarte LE. Aguirre-Dugua X, et al. Plants (Basel). 2023 Nov 27;12(23):3989. doi: 10.3390/plants12233989. Plants (Basel). 2023. PMID: 38068624 Free PMC article. - Insight on Rosaceae Family with Genome Sequencing and Functional Genomics Perspective.
Soundararajan P, Won SY, Kim JS. Soundararajan P, et al. Biomed Res Int. 2019 Feb 19;2019:7519687. doi: 10.1155/2019/7519687. eCollection 2019. Biomed Res Int. 2019. PMID: 30911547 Free PMC article. Review. - Genomics of the origin and evolution of Citrus.
Wu GA, Terol J, Ibanez V, López-García A, Pérez-Román E, Borredá C, Domingo C, Tadeo FR, Carbonell-Caballero J, Alonso R, Curk F, Du D, Ollitrault P, Roose ML, Dopazo J, Gmitter FG, Rokhsar DS, Talon M. Wu GA, et al. Nature. 2018 Feb 15;554(7692):311-316. doi: 10.1038/nature25447. Epub 2018 Feb 7. Nature. 2018. PMID: 29414943 - Population genetics of Mediterranean and Saharan olives: geographic patterns of differentiation and evidence for early generations of admixture.
Besnard G, El Bakkali A, Haouane H, Baali-Cherif D, Moukhli A, Khadari B. Besnard G, et al. Ann Bot. 2013 Nov;112(7):1293-302. doi: 10.1093/aob/mct196. Epub 2013 Sep 6. Ann Bot. 2013. PMID: 24013386 Free PMC article. - Insights into the effect of human civilization on Malus evolution and domestication.
Chen P, Li Z, Zhang D, Shen W, Xie Y, Zhang J, Jiang L, Li X, Shen X, Geng D, Wang L, Niu C, Bao C, Yan M, Li H, Li C, Yan Y, Zou Y, Micheletti D, Koot E, Ma F, Guan Q. Chen P, et al. Plant Biotechnol J. 2021 Nov;19(11):2206-2220. doi: 10.1111/pbi.13648. Epub 2021 Jul 7. Plant Biotechnol J. 2021. PMID: 34161653 Free PMC article.
References
- Diamond J. Guns, Germs, and Steel: The Fates of Human Societies. Norton, W. W. & Company, Inc. Sales; 1997.
- Zeder MA, Emshwiller E, Smith BD, Bradley DG. Documenting domestication: the intersection of genetics and archaeology. Trends Genet. 2006;22:139–155. - PubMed
- Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. - PubMed
- Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–848. - PubMed
- Wright SI, Gaut BS. Molecular population genetics and the search for adaptive evolution in plants. Mol Biol Evol. 2005;22:506–519. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources