Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence - PubMed (original) (raw)
Review
Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence
Taylor Heald-Sargent et al. Viruses. 2012 Apr.
Abstract
Coronavirus-cell entry programs involve virus-cell membrane fusions mediated by viral spike (S) proteins. Coronavirus S proteins acquire membrane fusion competence by receptor interactions, proteolysis, and acidification in endosomes. This review describes our current understanding of the S proteins, their interactions with and their responses to these entry triggers. We focus on receptors and proteases in prompting entry and highlight the type II transmembrane serine proteases (TTSPs) known to activate several virus fusion proteins. These and other proteases are essential cofactors permitting coronavirus infection, conceivably being in proximity to cell-surface receptors and thus poised to split entering spike proteins into the fragments that refold to mediate membrane fusion. The review concludes by noting how understanding of coronavirus entry informs antiviral therapies.
Keywords: membrane fusion; angiotensin converting enzyme 2; carcinoembryonic antigen; cathepsin; coronavirus; endocytosis; spike protein; transmembrane protease; viral pathogenesis; virus entry.
Figures
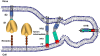
Figure 1
CoV virion and spike protein features. (A) Depiction of the virion. The virion core includes a helical ribonucleoprotein core consisting of a ~30 kilobase single-stranded RNA that is surrounded by nucleocapsid (N) proteins. The virion membrane is enriched with viral membrane proteins and includes a small number of envelope proteins. The spikes protrude about 20 nm from the limiting virion membrane; (B) Depiction of the S protein. A single S protein is depicted as a rectangle, from N- to C-terminus in left-to-right orientation. Relevant structural features are higlighted as follows: N-terminal receptor binding domain (N-RBD) in dark blue, with receptor binding motif (RBM) in yellow; C-RBD in brown, with RBM in yellow; cleavage sites (CS) 1 and 2, fusion peptide (FP) in red, heptad repeat (HR) regions 1 and 2 in green, with N and C-termini in yellow; transmembrane span ( TM ) depicted as membrane bilayer; cytoplasmic tail (CT) in light blue. The image is adjusted to the primary amino acid scale, which is 1255 residues for the SARS-CoV S protein. Note that the RBDs that are depicted come from different CoV S proteins; N-RBD from MHV and C-RBD from SARS; (C) Structure of the MHV N-RBD in complex with its CEACAM receptor (PDB 3R4D; reference [8]). The alpha carbon structure of the MHV N-RBD (in blue) is highlighted by RBMs (in yellow). The RBMs contact the N-terminal CEACAM receptor immunoglobulin domain (in purple); (D) Structure of the SARS C-RBD in complex with its ACE2 receptor (PDB 2AJF; reference [7]). The SARS C-RBD (in brown) is highlited by RBMs (in yellow). The RBMs contact a virus-binding hotspot [14] on the ACE2 receptor (in purple); (E) Structure of the postfusion HR1-HR2 bundle (PBD 1WYY; reference [15]). The HR1-HR2 bundle is depicted as alpha carbon tracings. Each of the three HR1-HR2 are a distinguished by color. To enhance the display, a single HR1-HR2 is extracted from the image and included below the 6-stranded bundle. This HR1-HR2 includes highlights (in yellow) depicting the N-terminal end of the HR1 and the C-terminal end of the HR2. The postfusion configuration is shown as it might appear relative to the fused membrane, when the fusion peptide (not shown) would extend into the fused membrane from the N-terminal end of HR1 and the HR2 would be held into the fused membrane by the TM span.
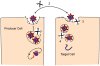
Figure 2
Schematic illustration of CoV S protein-mediated membrane fusion. The illustrations represent several steps of S protein conformational changes that may take place during membrane fusion. In the first step, receptor binding, pH reduction and/or S protein proteolysis induces dissociation of S1 from S2. This step is documented for some MHVs [29,30]. In the second step, the fusion peptide (FP) is intercalated into the host cell membrane. This is the fusion-intermediate stage. In the third stage, the part of the S protein nearest to the virus membrane refolds onto a heptad repeat 1 (HR1) core to form the six-helix bundle (6-HB), which is the final postfusion configuration of the S2 protein.
Figure 3
Proteolytic events during the CoV infection cycle. S cleavage events may take place during virus egress from producer cells (1), extracellular transit to target cells (2), on the target cell surface (3), and within the acidified endosome (4).
Similar articles
- Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease.
Bertram S, Glowacka I, Müller MA, Lavender H, Gnirss K, Nehlmeier I, Niemeyer D, He Y, Simmons G, Drosten C, Soilleux EJ, Jahn O, Steffen I, Pöhlmann S. Bertram S, et al. J Virol. 2011 Dec;85(24):13363-72. doi: 10.1128/JVI.05300-11. Epub 2011 Oct 12. J Virol. 2011. PMID: 21994442 Free PMC article. - Mechanisms of coronavirus cell entry mediated by the viral spike protein.
Belouzard S, Millet JK, Licitra BN, Whittaker GR. Belouzard S, et al. Viruses. 2012 Jun;4(6):1011-33. doi: 10.3390/v4061011. Epub 2012 Jun 20. Viruses. 2012. PMID: 22816037 Free PMC article. Review. - The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies.
Gierer S, Bertram S, Kaup F, Wrensch F, Heurich A, Krämer-Kühl A, Welsch K, Winkler M, Meyer B, Drosten C, Dittmer U, von Hahn T, Simmons G, Hofmann H, Pöhlmann S. Gierer S, et al. J Virol. 2013 May;87(10):5502-11. doi: 10.1128/JVI.00128-13. Epub 2013 Mar 6. J Virol. 2013. PMID: 23468491 Free PMC article. - Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response.
Glowacka I, Bertram S, Müller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, Niemeyer D, Schneider H, Drosten C, Pöhlmann S. Glowacka I, et al. J Virol. 2011 May;85(9):4122-34. doi: 10.1128/JVI.02232-10. Epub 2011 Feb 16. J Virol. 2011. PMID: 21325420 Free PMC article. - [Protease-dependent cell entry mechanism of coronaviruses].
Matsuyama S. Matsuyama S. Uirusu. 2011 Jun;61(1):109-16. doi: 10.2222/jsv.61.109. Uirusu. 2011. PMID: 21972562 Review. Japanese.
Cited by
- Emerging strategies on in silico drug development against COVID-19: challenges and opportunities.
Yadav M, Dhagat S, Eswari JS. Yadav M, et al. Eur J Pharm Sci. 2020 Dec 1;155:105522. doi: 10.1016/j.ejps.2020.105522. Epub 2020 Aug 20. Eur J Pharm Sci. 2020. PMID: 32827661 Free PMC article. Review. - Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+ endolysosomes is a rate-defining step.
Mingo RM, Simmons JA, Shoemaker CJ, Nelson EA, Schornberg KL, D'Souza RS, Casanova JE, White JM. Mingo RM, et al. J Virol. 2015 Mar;89(5):2931-43. doi: 10.1128/JVI.03398-14. Epub 2014 Dec 31. J Virol. 2015. PMID: 25552710 Free PMC article. - Contributions of human ACE2 and TMPRSS2 in determining host-pathogen interaction of COVID-19.
Senapati S, Banerjee P, Bhagavatula S, Kushwaha PP, Kumar S. Senapati S, et al. J Genet. 2021;100(1):12. doi: 10.1007/s12041-021-01262-w. J Genet. 2021. PMID: 33707363 Free PMC article. Review. - Proline-Proline Dyad in the Fusion Peptide of the Murine β-Coronavirus Spike Protein's S2 Domain Modulates Its Neuroglial Tropism.
Safiriyu AA, Mulchandani V, Anakkacheri MN, Pal D, Das Sarma J. Safiriyu AA, et al. Viruses. 2023 Jan 12;15(1):215. doi: 10.3390/v15010215. Viruses. 2023. PMID: 36680255 Free PMC article. - Swine Enteric Coronavirus: Diverse Pathogen-Host Interactions.
Yan Q, Liu X, Sun Y, Zeng W, Li Y, Zhao F, Wu K, Fan S, Zhao M, Chen J, Yi L. Yan Q, et al. Int J Mol Sci. 2022 Apr 2;23(7):3953. doi: 10.3390/ijms23073953. Int J Mol Sci. 2022. PMID: 35409315 Free PMC article. Review.
References
- Tooze J., Tooze S., Warren G. Replication of coronavirus mhv-a59 in sac- cells: Determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 1984;33:281–293. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources