Evolution and functional impact of rare coding variation from deep sequencing of human exomes - PubMed (original) (raw)
. 2012 Jul 6;337(6090):64-9.
doi: 10.1126/science.1219240. Epub 2012 May 17.
Abigail W Bigham, Timothy D O'Connor, Wenqing Fu, Eimear E Kenny, Simon Gravel, Sean McGee, Ron Do, Xiaoming Liu, Goo Jun, Hyun Min Kang, Daniel Jordan, Suzanne M Leal, Stacey Gabriel, Mark J Rieder, Goncalo Abecasis, David Altshuler, Deborah A Nickerson, Eric Boerwinkle, Shamil Sunyaev, Carlos D Bustamante, Michael J Bamshad, Joshua M Akey; Broad GO; Seattle GO; NHLBI Exome Sequencing Project
Affiliations
- PMID: 22604720
- PMCID: PMC3708544
- DOI: 10.1126/science.1219240
Evolution and functional impact of rare coding variation from deep sequencing of human exomes
Jacob A Tennessen et al. Science. 2012.
Abstract
As a first step toward understanding how rare variants contribute to risk for complex diseases, we sequenced 15,585 human protein-coding genes to an average median depth of 111× in 2440 individuals of European (n = 1351) and African (n = 1088) ancestry. We identified over 500,000 single-nucleotide variants (SNVs), the majority of which were rare (86% with a minor allele frequency less than 0.5%), previously unknown (82%), and population-specific (82%). On average, 2.3% of the 13,595 SNVs each person carried were predicted to affect protein function of ~313 genes per genome, and ~95.7% of SNVs predicted to be functionally important were rare. This excess of rare functional variants is due to the combined effects of explosive, recent accelerated population growth and weak purifying selection. Furthermore, we show that large sample sizes will be required to associate rare variants with complex traits.
Figures
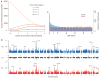
Fig. 1
Characteristics of protein-coding variation in humans. (A) Number of nonsynonymous SNVs predicted to be functionally important as a function of seven different methods (18). (B) Distributions of π across the exome in AAs (blue) and EAs (red). The value of π for each gene is shown as a vertical line. The middle section shows the difference in diversity between EA and AA (Δπ = πEA − πAA), scaled between 0 and 1. (C) Distributions of the proportion of total diversity, π, attributable to SNVs with different MAFs in the EA and AA samples. The x axis is binned in increments of 0.5%.
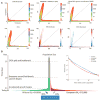
Fig. 2
Deep sequencing reveals increases of recent population size. (A) Joint SFS predicted from different demographic models (top) compared with the observed data (bottom), displaying allele counts between 0 and 100 chromosomes. The three models are (left) an OOA model without admixture derived from the 1000 Genomes data, (middle) the same model with the AA panel modeled as an 80%:20% admixture between African and European lineages, and (right) the same model further modified to account for recent growth acceleration. Anscombe residuals are displayed, with regions showing more variants than predicted by the model in blue and less in red. Bins with expected counts <1 are displayed as white in all graphs. (B) Schematic representation (not to scale) of the inferred demographic model and parameters (18). kya, thousand years ago. (Inset) Comparison of the observed SFS to that predicted by the demographic model incorporating recent accelerated growth.
Fig. 3
Signatures of purifying selection in protein-coding SNVs. (A) Relationship between the evidence that a variant is functionally important and MAF for four different methods. (B) Relationship between the proportion of putatively functional variants and MAF for the same predictions as in (A). (C) Comparison of the number of rare SNVs (orange) and enrichmentofrareornon-synonymous SNVs (brown) located in different protein structural categories [P values were calculated by a permutation test (18)]. (D) Relationship between average change of w score of synonymous variants and DAF.
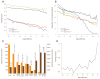
Fig. 4
Power of rare variant association mapping and personal genomics characteristics of protein-coding SNVs. (A) Distribution of gene-specific estimates of power to map causal rare variants across 12,000 protein-coding genes with at least three SNVs in the EA (red) or AA (blue) samples. Power varied widely across loci, and <5% of genes (beige) achieve 80% power even when relatively strong effects (OR = 5) are modeled. (B) Average number (points) and range (vertical lines) of synonymous, missense, splice site, and nonsense SNVs. (C) Average proportion of SNVs per individual that are rare (MAF ≤ 0.5%), intermediate (0.5% < MAF < 5%), or common (MAF ≥ 5%) in the population from which they were sampled. The proportions of rare and intermediate frequency variants per individual are significantly higher (Wilcoxon-rank sum test; P < 10−15) for putatively functional SNVs. (D) Violin plots showing the distribution of number of functional SNVs, number of functional singletons, and proportion of functional SNVs per individual in the EA and AA samples. Darker and lighter shaded plots correspond to conservative and more liberal definitions of functional variation, respectively.
Comment in
- Genetics. Human genetic variation, shared and private.
Casals F, Bertranpetit J. Casals F, et al. Science. 2012 Jul 6;337(6090):39-40. doi: 10.1126/science.1224528. Science. 2012. PMID: 22767915 No abstract available.
Similar articles
- An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people.
Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Shen J, Tang Z, Bacanu SA, Fraser D, Warren L, Aponte J, Zawistowski M, Liu X, Zhang H, Zhang Y, Li J, Li Y, Li L, Woollard P, Topp S, Hall MD, Nangle K, Wang J, Abecasis G, Cardon LR, Zöllner S, Whittaker JC, Chissoe SL, Novembre J, Mooser V. Nelson MR, et al. Science. 2012 Jul 6;337(6090):100-4. doi: 10.1126/science.1217876. Epub 2012 May 17. Science. 2012. PMID: 22604722 Free PMC article. - Genetics. Human genetic variation, shared and private.
Casals F, Bertranpetit J. Casals F, et al. Science. 2012 Jul 6;337(6090):39-40. doi: 10.1126/science.1224528. Science. 2012. PMID: 22767915 No abstract available. - Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants.
Fu W, O'Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ; NHLBI Exome Sequencing Project; Akey JM. Fu W, et al. Nature. 2013 Jan 10;493(7431):216-20. doi: 10.1038/nature11690. Epub 2012 Nov 28. Nature. 2013. PMID: 23201682 Free PMC article. - Explosive genetic evidence for explosive human population growth.
Gao F, Keinan A. Gao F, et al. Curr Opin Genet Dev. 2016 Dec;41:130-139. doi: 10.1016/j.gde.2016.09.002. Epub 2016 Oct 4. Curr Opin Genet Dev. 2016. PMID: 27710906 Free PMC article. Review. - Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data.
Cooper GM, Shendure J. Cooper GM, et al. Nat Rev Genet. 2011 Aug 18;12(9):628-40. doi: 10.1038/nrg3046. Nat Rev Genet. 2011. PMID: 21850043 Review.
Cited by
- The Use of Next-Generation Sequencing in Personalized Medicine.
Popova L, Carabetta VJ. Popova L, et al. Methods Mol Biol. 2025;2866:287-315. doi: 10.1007/978-1-0716-4192-7_16. Methods Mol Biol. 2025. PMID: 39546209 Review. - Leveraging ancient DNA to uncover signals of natural selection in Europe lost due to admixture or drift.
Pandey D, Harris M, Garud NR, Narasimhan VM. Pandey D, et al. Nat Commun. 2024 Nov 12;15(1):9772. doi: 10.1038/s41467-024-53852-8. Nat Commun. 2024. PMID: 39532856 Free PMC article. - Improving long-term kidney allograft survival by rethinking HLA compatibility: from molecular matching to non-HLA genes.
Mattoo A, Jaffe IS, Keating B, Montgomery RA, Mangiola M. Mattoo A, et al. Front Genet. 2024 Oct 2;15:1442018. doi: 10.3389/fgene.2024.1442018. eCollection 2024. Front Genet. 2024. PMID: 39415982 Free PMC article. Review. - Deep Ancestral Introgressions between Ovine Species Shape Sheep Genomes via Argali-Mediated Gene Flow.
Lv FH, Wang DF, Zhao SY, Lv XY, Sun W, Nielsen R, Li MH. Lv FH, et al. Mol Biol Evol. 2024 Nov 1;41(11):msae212. doi: 10.1093/molbev/msae212. Mol Biol Evol. 2024. PMID: 39404100 Free PMC article. - Constraining models of dominance for nonsynonymous mutations in the human genome.
Kyriazis CC, Lohmueller KE. Kyriazis CC, et al. PLoS Genet. 2024 Sep 20;20(9):e1011198. doi: 10.1371/journal.pgen.1011198. eCollection 2024 Sep. PLoS Genet. 2024. PMID: 39302992 Free PMC article.
References
- Bamshad MJ, et al. Nat Rev Genet. 2011;12:745. - PubMed
- International HapMap Consortium. Nature. 2005;437:1299. - PubMed
Publication types
MeSH terms
Grants and funding
- RC2 HL102923/HL/NHLBI NIH HHS/United States
- RC2 HL102926/HL/NHLBI NIH HHS/United States
- RC2 HL-102926/HL/NHLBI NIH HHS/United States
- U01 HG006513/HG/NHGRI NIH HHS/United States
- RC2 HL-102923/HL/NHLBI NIH HHS/United States
- R01 HG003229/HG/NHGRI NIH HHS/United States
- RC2 HL-102925/HL/NHLBI NIH HHS/United States
- RC2 HL103010/HL/NHLBI NIH HHS/United States
- RC2 HL-102924/HL/NHLBI NIH HHS/United States
- RC2 HL102924/HL/NHLBI NIH HHS/United States
- RC2 HL-103010/HL/NHLBI NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials