Mutational processes molding the genomes of 21 breast cancers - PubMed (original) (raw)
. 2012 May 25;149(5):979-93.
doi: 10.1016/j.cell.2012.04.024. Epub 2012 May 17.
Ludmil B Alexandrov, David C Wedge, Peter Van Loo, Christopher D Greenman, Keiran Raine, David Jones, Jonathan Hinton, John Marshall, Lucy A Stebbings, Andrew Menzies, Sancha Martin, Kenric Leung, Lina Chen, Catherine Leroy, Manasa Ramakrishna, Richard Rance, King Wai Lau, Laura J Mudie, Ignacio Varela, David J McBride, Graham R Bignell, Susanna L Cooke, Adam Shlien, John Gamble, Ian Whitmore, Mark Maddison, Patrick S Tarpey, Helen R Davies, Elli Papaemmanuil, Philip J Stephens, Stuart McLaren, Adam P Butler, Jon W Teague, Göran Jönsson, Judy E Garber, Daniel Silver, Penelope Miron, Aquila Fatima, Sandrine Boyault, Anita Langerød, Andrew Tutt, John W M Martens, Samuel A J R Aparicio, Åke Borg, Anne Vincent Salomon, Gilles Thomas, Anne-Lise Børresen-Dale, Andrea L Richardson, Michael S Neuberger, P Andrew Futreal, Peter J Campbell, Michael R Stratton; Breast Cancer Working Group of the International Cancer Genome Consortium
Affiliations
- PMID: 22608084
- PMCID: PMC3414841
- DOI: 10.1016/j.cell.2012.04.024
Mutational processes molding the genomes of 21 breast cancers
Serena Nik-Zainal et al. Cell. 2012.
Abstract
All cancers carry somatic mutations. The patterns of mutation in cancer genomes reflect the DNA damage and repair processes to which cancer cells and their precursors have been exposed. To explore these mechanisms further, we generated catalogs of somatic mutation from 21 breast cancers and applied mathematical methods to extract mutational signatures of the underlying processes. Multiple distinct single- and double-nucleotide substitution signatures were discernible. Cancers with BRCA1 or BRCA2 mutations exhibited a characteristic combination of substitution mutation signatures and a distinctive profile of deletions. Complex relationships between somatic mutation prevalence and transcription were detected. A remarkable phenomenon of localized hypermutation, termed "kataegis," was observed. Regions of kataegis differed between cancers but usually colocalized with somatic rearrangements. Base substitutions in these regions were almost exclusively of cytosine at TpC dinucleotides. The mechanisms underlying most of these mutational signatures are unknown. However, a role for the APOBEC family of cytidine deaminases is proposed.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
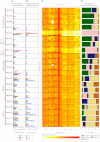
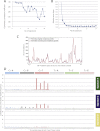
Somatic Mutation Profiles of 21 Breast Cancers, Related to Table S1 Breast cancers grouped according to subtype on the far left. (A) Base substitution mutation spectra. ∗Ultra-deep sequenced PD4120a has an alternative scale on the x axis (0 to 45,000). (B) Mutation spectra of double substitutions from all 21 samples. (C) Genomic heatmap constructed from counts of each mutation-type at each mutation context corrected for the frequency of each trinucleotide in the reference genome. Log-transformed values of these ratios have been plotted in the heatmap. The 5′ base to each mutated base is shown on the vertical axis and 3′ base on the horizontal axis. Heatmap scale at the bottom. (D) Proportion of the total substitutions contributed by each of the five mutational signatures, as identified by NMF analysis, for all 21 cancer genomes.
Figure 2
Five Mutational Signatures Extracted by NMF in 21 Breast Cancers, Related to Figure S1 (A) Fraction of contribution of each mutation-type at each context for the five mutational signatures identified by NMF analysis. The major components contributing to each signature are highlighted with arrows. (B) Cluster dendrogram generated by unsupervised hierarchical clustering based on contributions of the five mutational signatures identified by NMF to the 21 breast cancer genomes.
Figure 3
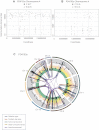
Kataegis, Regional Hypermutation of Base Substitutions, Related to Figure S2 (A) Rainfall plot of PD4107a. Mutations are ordered on the x axis from the first variant on the short arm of chromosome 1 to the last variant on the long arm of chromosome X and are colored according to mutation-type. The distance between each mutation and the one prior to it (the intermutation distance) is plotted on the vertical axis on a log scale. Most mutations in this genome have an intermutation distance of ∼105 bp to ∼106 bp. Mutations in a region of hypermutation present as a cluster of lower intermutation distances. (B) Rainfall plot for PD4103a demonstrating kataegis occurring at multiple loci through the genome. (C) Rainfall plot for PD4085a, showing no kataegis. (D) Plots of flanking sequence of all C>X mutations and C>X mutations within the regions of kataegis in PD4107a. Mutated base is at position 0 with ten bases of flanking sequence provided, demonstrating a strong preference for T at the −1 position.
Figure 4
Rainfall Plot for Chromosome 6 of PD4107a (A) The x axis shows the genomic coordinates of the mutations. Rearrangements are presented as brown triangles (rearrs is an abbreviation for rearrangements). The region of kataegis is highlighted at increasing resolution to demonstrate microclusters within the macrocluster. The processive nature of C>T mutations at TpC context occurring in cis is seen in the lowest panel (G-browse image). (B) Alternating processivity of kataegis in PD4107a. Long regions of C>T mutations are interspersed with regions of G>A mutations. (C) Kataegis occurs with a variety of rearrangement architectures. Thick top line shows the copy number segments for the region of chromosome 6 of PD4107a. Point mutations are shown in lower panel as black points. x axis reflecting genomic position and y axis represents variant allele fraction. The proportions of reads derived from contaminating normal cells are depicted in gray and the fraction coming from each of the copies of that segment in the tumor cells are depicted by the multiple bars from green to yellow to pink to white. Early mutations will be found relatively higher up these bars, whereas late ones will be seen down the bottom of the variant allele fraction. Grey vertical lines represent rearrangements. Interconnecting lines indicate intrachromosomal rearrangements. On a macroscopic scale, this demonstrates how kataegis can be associated with chromothripsis (within region 130–135 Mb) as well as other rearrangement architectures.
Figure 5
Processivity and Complex Colocalization of Rearrangement Architecture with Kataegis in PD4103a (A) Stretches of C>T alternate with stretches of G>A on chromosome 4 in PD4103a. (B) Alternating C>G and G>C mutation on the same chromosome in PD4103a. (C) The complex web of rearrangements involving 8 chromosomes in PD4103a colocalizing with kataegis.
Figure 6
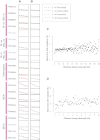
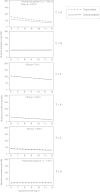
Relationship between Mutation Prevalence and Transcription and/or Expression, Related to Figure S3 Mutation prevalence is expressed as the number of mutations per Mb from 0 to 2 per Mb on the vertical axis. Log 2 expression levels range from 6 to 12 on the horizontal axis. Lines are fitted curves to the data for A and B. (A) C>A mutations; and (B) T>A mutations. Breast cancer samples without expression data are shown in gray. (C) Effect of distance from transcription start site on mutation prevalence. Each dot represents a 1 kb bin at increasing distances from all transcription start sites (TSS) up to 200 kb. The y axis shows the percentage of genes in each bin carrying a somatic mutation. The mutation prevalence increases as distance increases from the TSS. (D) This is particularly marked in the first 1 kb after the TSS. Each dot represents a 100 bp bin.
Figure 7
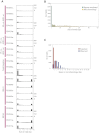
Somatic Mutation Profile of Indels (A) The x axis shows indel size from 1–10 and all larger indels between 11-50 bp in size grouped in a single bin. The y axis shows the number in each genome from 0–300. (B) Frequency of indels by indel size. This demonstrates how repeat-mediated indels are usually of smaller size. From a Kolmogorov-Smirnov (K-S) test, the distribution of indel lengths for repeats and microhomologies is significantly different (p < 2.2 × 10−16). (C) Observed number of bases involved in microhomology at junction of indels versus expected number of bases if microhomology occurred simply by chance.
Figure S1
Selection of the Optimal Number of Signatures via the NMF Model Selection Framework, Related to Figure 2 (A) The x axis depicts the number of signatures, whereas the y axis shows the cophenetic coefficient. As an indicator of stable reproducibility, the cophenetic correlation coefficient is at its highest points at between 2 and 6 processes. Given that there are no further peaks after 6 for this data set, the number of signatures recognized by the NMF algorithm here is up to six. (B) The error in reconstruction for each number of potential signatures, k, showed a marked reduction in the slope of the reconstruction error until k = 5, suggesting that the model was stable at five mutational signatures. (C) A typical comparison between the reconstructed and original mutation profile demonstrating how well the extracted signatures and their exposures describe the original data for five signatures. (D) Signatures A,C and D with contributions from each of the 96 trinucleotides corrected for the frequency of trinucleotides in the genome. This form of representation highlights the contrast between Signature A and C, as well as demonstrates the differences between Signatures C and D. Note the absence of C > T transitions at Xp
C
pG in Signature D.
Figure S2
Rainfall Plots for 18 Genomes, Related to Figure 3 PD4115a, PD4116a, PD3904a, PD3945a, PD4005a and PD4006a show an excess of mutations of intermutation distance of 1bp, in-keeping with the observed excess of double substitutions in these genomes. Subtle regions of kataegis are present in many samples (PD4199a, PD4192a, PD4198a, PD4248a, PD4116a, PD3904a, PD4005a and PD4006a). Intermutation distance (bp) is presented on the vertical axis and mutation number is presented on the horizontal axis.
Figure S3
Relationship between Mutation Prevalence, Transcription and Gene Expression, Related to Figure 6 Overall effect of transcription and gene expression on mutation prevalence by mutation type. p values of significance are provided for each mutation-type if a strong effect was seen in either strand bias and/or relationship with expression. Mutation prevalence is expressed as the number of mutations per Mb from 0 to 2 per Mb on the vertical axis. Log 2 expression levels range from 6 to 12 on the horizontal axis. Lines are fitted curves to the data for A and B.
Comment in
- Tumor archaeology reveals that mutations love company.
Setlur SR, Lee C. Setlur SR, et al. Cell. 2012 May 25;149(5):959-61. doi: 10.1016/j.cell.2012.05.010. Cell. 2012. PMID: 22632962 No abstract available.
Similar articles
- DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis.
Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. Taylor BJ, et al. Elife. 2013 Apr 16;2:e00534. doi: 10.7554/eLife.00534. Elife. 2013. PMID: 23599896 Free PMC article. - Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer.
Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, Davies HR, Knappskog S, Martin S, Papaemmanuil E, Ramakrishna M, Shlien A, Simonic I, Xue Y, Tyler-Smith C, Campbell PJ, Stratton MR. Nik-Zainal S, et al. Nat Genet. 2014 May;46(5):487-91. doi: 10.1038/ng.2955. Epub 2014 Apr 13. Nat Genet. 2014. PMID: 24728294 Free PMC article. - Integrative genomic analysis identifies associations of molecular alterations to APOBEC and BRCA1/2 mutational signatures in breast cancer.
Trevino V. Trevino V. Mol Genet Genomic Med. 2019 Aug;7(8):e810. doi: 10.1002/mgg3.810. Epub 2019 Jul 11. Mol Genet Genomic Med. 2019. PMID: 31294536 Free PMC article. - Deaminase-Driven Reverse Transcription Mutagenesis in Oncogenesis: Critical Analysis of Transcriptional Strand Asymmetries of Single Base Substitution Signatures.
Steele EJ, Lindley RA. Steele EJ, et al. Int J Mol Sci. 2025 Jan 24;26(3):989. doi: 10.3390/ijms26030989. Int J Mol Sci. 2025. PMID: 39940758 Free PMC article. Review. - Mutational spectra and mutational signatures: Insights into cancer aetiology and mechanisms of DNA damage and repair.
Phillips DH. Phillips DH. DNA Repair (Amst). 2018 Nov;71:6-11. doi: 10.1016/j.dnarep.2018.08.003. Epub 2018 Aug 24. DNA Repair (Amst). 2018. PMID: 30236628 Free PMC article. Review.
Cited by
- MiST: a new approach to variant detection in deep sequencing datasets.
Subramanian S, Di Pierro V, Shah H, Jayaprakash AD, Weisberger I, Shim J, George A, Gelb BD, Sachidanandam R. Subramanian S, et al. Nucleic Acids Res. 2013 Sep;41(16):e154. doi: 10.1093/nar/gkt551. Epub 2013 Jul 4. Nucleic Acids Res. 2013. PMID: 23828039 Free PMC article. - Efficient deamination of 5-methylcytidine and 5-substituted cytidine residues in DNA by human APOBEC3A cytidine deaminase.
Suspène R, Aynaud MM, Vartanian JP, Wain-Hobson S. Suspène R, et al. PLoS One. 2013 Jun 20;8(6):e63461. doi: 10.1371/journal.pone.0063461. Print 2013. PLoS One. 2013. PMID: 23840298 Free PMC article. - Mutational Signature and Transcriptomic Classification Analyses as the Decisive Diagnostic Tools for a Cancer of Unknown Primary.
Bagge RO, Demir A, Karlsson J, Alaei-Mahabadi B, Einarsdottir BO, Jespersen H, Lindberg MF, Muth A, Nilsson LM, Persson M, Svensson JB, Söderberg EMV, de Krijger RR, Nilsson O, Larsson E, Stenman G, Nilsson JA. Bagge RO, et al. JCO Precis Oncol. 2018 Jul 30;2:PO.18.00002. doi: 10.1200/PO.18.00002. eCollection 2018. JCO Precis Oncol. 2018. PMID: 32913988 Free PMC article. - Characterization of the mechanism by which the RB/E2F pathway controls expression of the cancer genomic DNA deaminase APOBEC3B.
Roelofs PA, Goh CY, Chua BH, Jarvis MC, Stewart TA, McCann JL, McDougle RM, Carpenter MA, Martens JW, Span PN, Kappei D, Harris RS. Roelofs PA, et al. Elife. 2020 Sep 28;9:e61287. doi: 10.7554/eLife.61287. Elife. 2020. PMID: 32985974 Free PMC article. - Defining relative mutational difficulty to understand cancer formation.
Shan L, Yu J, He Z, Chen S, Liu M, Ding H, Xu L, Zhao J, Yang A, Jiang H. Shan L, et al. Cell Discov. 2020 Jul 21;6:48. doi: 10.1038/s41421-020-0177-8. eCollection 2020. Cell Discov. 2020. PMID: 32704382 Free PMC article.
References
- Berry M.W., Browne M., Langville A.N., Pauca V.P., Plemmons R.J. Algorithms and applications for approximate nonnegative matrix factorization. Comput. Stat. Data Anal. 2007;52:155–173.
Supplemental References
- Campbell, P.J., Stephens, P.J., Pleasance, E.D., O'Meara, S., Li, H., Santarius, T., Stebbings, L.A., Leroy, C., Edkins, S., Hardy, C., et al. (2008). Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 40, 722–729. - PMC - PubMed
- Lee, D.D., and Seung, H.S. (1999). Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA089393/CA/NCI NIH HHS/United States
- MC_U105178806/MRC_/Medical Research Council/United Kingdom
- P50 CA089393/CA/NCI NIH HHS/United States
- DH_/Department of Health/United Kingdom
- 088340/WT_/Wellcome Trust/United Kingdom
- WT088340MA/WT_/Wellcome Trust/United Kingdom
- 098051/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous