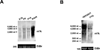Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons - PubMed (original) (raw)
Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons
Kate D Meyer et al. Cell. 2012.
Abstract
Methylation of the N(6) position of adenosine (m(6)A) is a posttranscriptional modification of RNA with poorly understood prevalence and physiological relevance. The recent discovery that FTO, an obesity risk gene, encodes an m(6)A demethylase implicates m(6)A as an important regulator of physiological processes. Here, we present a method for transcriptome-wide m(6)A localization, which combines m(6)A-specific methylated RNA immunoprecipitation with next-generation sequencing (MeRIP-Seq). We use this method to identify mRNAs of 7,676 mammalian genes that contain m(6)A, indicating that m(6)A is a common base modification of mRNA. The m(6)A modification exhibits tissue-specific regulation and is markedly increased throughout brain development. We find that m(6)A sites are enriched near stop codons and in 3' UTRs, and we uncover an association between m(6)A residues and microRNA-binding sites within 3' UTRs. These findings provide a resource for identifying transcripts that are substrates for adenosine methylation and reveal insights into the epigenetic regulation of the mammalian transcriptome.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Figure 1. Specificity and Sensitivity of m6A-Specific Antibody
A. Dot blot analysis demonstrates antibody specificity for m6A. Increasing amounts of an oligonucleotide containing either m6A or unmodified adenosine were spotted onto a membrane and probed with the m6A antibody. While increased m6A immunoreactivity is observed in the presence of increasing concentrations of the m6A oligonucleotide (top), only background levels of immunoreactivity are observed at the highest concentrations of the A oligonucleotide (bottom). Blots shown are representative of results from three experiments. B. Competition dot blot assays were performed on membranes spotted with 100 ng of m6A-containing oligonucleotide. Antibody binding to the m6A oligonucleotide is attenuated by pre-incubation with increasing amounts of m6A-containing competitor RNA (top), but not with RNA containing unmodified adenosine (bottom). Amount of competitor RNA used (left to right): 0 ng (0 nM), 10 ng (0.1 nM), 100 ng (1.1 nM), 1 µg (11.2 nM). Blots shown are representative of results from four experiments. C. Competition dot blot assays were performed as in (B). Antibody was pre-incubated with increasing amounts of _N_6-methyladenosine triphosphate (_N_6-MeATP), adenosine triphosphate (ATP), _N_1-methyladenosine triphosphate (_N_1-MeATP), or 2’-_O_-methyladenosine triphosphate (2’-_O_-MeATP). Only _N_6-MeATP is able to compete with antibody binding. Concentration of competitor nucleotide used (left to right): 0 µM, 1 µM, 2 µM, 4 µM. Blots shown are representative of results from three experiments. D. Detection of m6A in cellular DNA. Genomic DNA isolated from dam+ (containing m6A) or dam− (lacking m6A) E. coli was sheared and subjected to immunoblotting with the anti-m6A antibody. Although 1.5 times as much DNA from dam− E. coli was loaded (left panel), the antibody only recognizes the m6A present in DNA from dam+ E. coli (right panel). Blot shown is representative of results from three experiments. See also Figure S2.
Figure 2. Distribution and Dynamic Cellular Regulation of m6A in RNA
A. Widespread distribution of m6A levels in a variety of tissues. Total RNA isolated from mouse brain, heart, lung, liver, and kidney (top) was subjected to m6A immunoblot analysis. Ethidium bromide staining of the 28S rRNA is shown as a loading control (bottom). B. Quantification of m6A abundance within various tissues. Quantification of m6A immunoreactivity in (A) was measured by densitometry and normalized to the intensity of the corresponding 28S rRNA band for each tissue (n = 3; data are presented as mean ± SEM). C. m6A is enriched within mRNAs. Oligo(dT) Dynabeads were used to isolate poly(A) RNA from total mouse brain RNA, and the unbound “flow-through” RNA was saved as the poly(A)-depleted fraction. Equal amounts of total RNA, poly(A) RNA, and poly(A)-depleted RNA were then subjected to m6A immunoblot analysis (top). Ethidium bromide staining of 28S rRNA is shown as a loading control (bottom). Intense m6A immunoreactivity is observed in the poly(A) RNA fraction, consistent with high levels of m6A within mRNAs. D. Depletion of poly(A) tails from mRNA does not reduce levels of m6A in mRNA. Poly(A) RNA was isolated from total mouse brain RNA using oligo(dT) Dynabeads. Half the sample was then subjected to poly(A) tail depletion by hybridizing to oligo(dT) primers and digestion with RNase H. Immunoblot analysis with the m6A antibody (top panel) shows that levels of m6A in poly(A) RNA (left) and poly(A) tail-depleted RNA (right) are comparable. Removal of poly(A) tails was confirmed using 3’RACE and RTPCR to detect β-actin; no product is detected in the tail-depleted sample when oligo(dT) primers are used for cDNA synthesis (middle panel). As a control, use of random hexamers successfully generates a product in both samples (bottom panel). See also Figure S1.
Figure 3. Regulation of m6A Levels in Cells and During Development
A. Ontogeny of m6A abundance throughout brain development. Total RNA was isolated from mouse brain at embryonic day 18 (E18), postnatal day 0 (P0), postnatal day 14 (P14), and adulthood, then subjected to immunoblot analysis to detect m6A-containing transcripts. Ethidium bromide staining of 28S rRNA bands is shown as a loading control. B. FTO demethylates a wide range of cellular transcripts. FTO was expressed in HEK293T cells for 48h, and cellular RNA was subjected to immunoblot analysis to detect m6A. See also Figure S1.
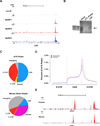
Figure 4. Outline of MeRIP-Seq Protocol and Distribution of Sequencing Reads
A. Schematic representation of MeRIP-Seq. Total RNA is subjected to RiboMinus treatment to remove rRNA species. RNAs containing m6A are then immunoprecipitated by mixing the RNA with m6A antibody-coupled Dynabeads. m6A-containing RNAs are then eluted from the antibody-coupled beads and subjected to a second round of m6A immunoprecipitation. The resulting RNA pool, which is highly enriched for m6A-containing RNAs, is then subjected to next-generation sequencing. B. Schematic of sequencing reads and their alignment to locations in the genome surrounding an m6A site. Top: an mRNA that contains a single m6A residue along its length. Middle: individual 100 nt-wide mRNA fragments which are isolated following m6A immunoprecipitation, each of which contains the same m6A residue from the mRNA depicted above. Bottom: histogram showing predicted frequency of MeRIP-Seq reads obtained by sequencing individual immunoprecipitated fragments. Read frequency is predicted to increase with closer proximity to the m6A site, forming a “peak” which is roughly 200 nt wide at its base and 100 nt wide at its midpoint. C. Sequencing reads from MeRIP-Seq converge over m6A sites. Representative UCSC Genome Browser plot from MeRIP-Seq data which demonstrates typical read frequency peak formation surrounding a site of m6A (shown here is the 3’ UTR of Pax6). Peak height is displayed as reads per base per million mapped reads (BPM). See also Figures S2, S3.
Figure 5. Validation of m6A Targets and Characteristics of m6A Localization
A. Different sequencing platforms and antibodies result in similar m6A profiles. UCSC Genome Browser tracks displaying read clusters from three MeRIP-Seq replicates (MeRIP1, MeRIP2, and MeRIP3) are shown along the length of the Ldlr transcript. The upper-most track (non-IP) represents the non-immunoprecipitated control sample. B. Validation of m6A-containing mRNA identified with MeRIP-Seq. Hybridization-based RNA pulldown was used to isolate Ldlr mRNA from total brain RNA, followed by confirmation of m6A presence (arrow) by immunoblot analysis with anti-m6A. A control sample using a non-specific probe of equal size (Control Probe) was run in parallel. Total mouse brain RNA (Input RNA) is shown as a reference for m6A labeling. C. Transcriptome-wide distribution of m6A peaks. Pie charts showing the percentage of m6A peaks (top) and non-IP sample reads (bottom) within distinct RNA sequence types. m6A is highly enriched in 3’ UTRs and CDSs compared to the distribution of reads in the non-IP samples. D. Distribution of m6A peaks across the length of mRNA transcripts. 5’ UTRs, CDSs, and 3’ UTRs of RefSeq mRNAs were individually binned into regions spanning 1% of their total length, and the percentage of m6A peaks that fall within each bin was determined. The moving averages of mouse brain peaks percentage (red) and HEK293T peak percentage (blue) are shown. E. Highly similar m6A peak distribution is observed within many human and mouse transcripts. UCSC Genome Browser plots showing MeRIP-Seq read clusters in the representative transcript SREK1. MeRIP-Seq reads cluster at the same distinct regions of SREK1 in both HEK293T cell RNA (top) and mouse brain RNA (bottom). See also Figures S4 – S7, Tables S1-S6.
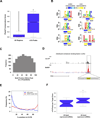
Figure 6. MeRIP-Seq Reveals Features of m6A in mRNA
A. Phylogenetic conservation of m6A peaks. PhyloP scores of m6A peak regions were compared to those of randomly shuffled regions throughout gene exons. There was a significantly higher median conservation score (K-S test, * p ≤ 2.2e−16) in m6A peaks (0.578) than in the random regions (0.023). B. Sequence motifs identified within m6A peaks. The motif G[AG]ACU and variants thereof ([AC]GAC[GU], GGAC, [AU][CG]G[AG]AC, and UGAC) was highly enriched in m6A peaks. Additionally, one U-rich motif (bottom right) was identified as being significantly underrepresented within m6A peaks. Color bars under each motif indicate the degree of underrepresentation (blue) or overrepresentation (yellow) within regions of m6A peaks in the non-IP control sample (CNTL) and the MeRIP sample (MeRIP). C. m6A motif sequences frequently lie near the center of m6A peaks. Shown is a plot of the cumulative distribution of m6A motif positions within m6A peaks containing a single motif. Motifs cluster in the center of peaks, suggesting that the methylated adenosines in these motifs account for the m6A peaks identified in MeRIP-Seq. D. Example of a m6A motif sequence near the center of a peak. UCSC Genome Browser plot containing tracks for MeRIP-Seq reads (red) and non-IP control reads (black) at the Ilf2 locus. The m6A peak within the Ilf2 3’ UTR contains a single m6A motif identified in (B). The sequence of this motif (highlighted in yellow) is located at the center of the m6A peak. E. Distribution of m6A peaks and miRNA target sites within 3’ UTRs. The frequency of m6A peaks (blue) and miRNA target sites (red) along the length of 3’ UTRs is shown. F. Association between 3’ UTR methylation and miRNA abundance. The 25 most abundant miRNAs in brain have a significantly greater percentage of m6A peaks within their target mRNA 3’ UTRs than do the 25 most weakly expressed brain miRNAs (*p<0.05, Wilcoxon test). The error bars in A and F indicate the highest and lowest values, and the box boundaries denote the 1st quartile, median, and 3rd quartile. See also Figures S3, S6, and S7.
Comment in
- Implications of widespread covalent modification of mRNA.
Cooper TA. Cooper TA. Circ Res. 2012 Dec 7;111(12):1491-3. doi: 10.1161/CIRCRESAHA.112.281071. Circ Res. 2012. PMID: 23223930 No abstract available. - An epigenetics gold rush: new controls for gene expression.
Willyard C. Willyard C. Nature. 2017 Feb 22;542(7642):406-408. doi: 10.1038/542406a. Nature. 2017. PMID: 28230146 No abstract available.
Similar articles
- Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome.
Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. Li X, et al. Nat Chem Biol. 2016 May;12(5):311-6. doi: 10.1038/nchembio.2040. Epub 2016 Feb 10. Nat Chem Biol. 2016. PMID: 26863410 - Transcriptome-wide N 6 -methyladenosine methylome profiling of porcine muscle and adipose tissues reveals a potential mechanism for transcriptional regulation and differential methylation pattern.
Tao X, Chen J, Jiang Y, Wei Y, Chen Y, Xu H, Zhu L, Tang G, Li M, Jiang A, Shuai S, Bai L, Liu H, Ma J, Jin L, Wen A, Wang Q, Zhu G, Xie M, Wu J, He T, Huang C, Gao X, Li X. Tao X, et al. BMC Genomics. 2017 Apr 28;18(1):336. doi: 10.1186/s12864-017-3719-1. BMC Genomics. 2017. PMID: 28454518 Free PMC article. - Transcriptome-wide analysis reveals spatial correlation between N6-methyladenosine and binding sites of microRNAs and RNA-binding proteins.
Das Mandal S, Ray PS. Das Mandal S, et al. Genomics. 2021 Jan;113(1 Pt 1):205-216. doi: 10.1016/j.ygeno.2020.12.027. Epub 2020 Dec 16. Genomics. 2021. PMID: 33340693 - Regulatory Role of N6 -methyladenosine (m6 A) Methylation in RNA Processing and Human Diseases.
Wei W, Ji X, Guo X, Ji S. Wei W, et al. J Cell Biochem. 2017 Sep;118(9):2534-2543. doi: 10.1002/jcb.25967. Epub 2017 May 15. J Cell Biochem. 2017. PMID: 28256005 Review. - N6-methyl-adenosine modification in messenger and long non-coding RNA.
Pan T. Pan T. Trends Biochem Sci. 2013 Apr;38(4):204-9. doi: 10.1016/j.tibs.2012.12.006. Epub 2013 Jan 19. Trends Biochem Sci. 2013. PMID: 23337769 Free PMC article. Review.
Cited by
- Genetically encoded chemical crosslinking of RNA in vivo.
Sun W, Wang N, Liu H, Yu B, Jin L, Ren X, Shen Y, Wang L. Sun W, et al. Nat Chem. 2023 Jan;15(1):21-32. doi: 10.1038/s41557-022-01038-4. Epub 2022 Oct 6. Nat Chem. 2023. PMID: 36202986 Free PMC article. - Transcriptome-Wide Study of mRNAs and lncRNAs Modified by m6A RNA Methylation in the Longissimus Dorsi Muscle Development of Cattle-Yak.
Huang C, Dai R, Meng G, Dingkao R, Wang X, Ren W, Ma X, Wu X, Chu M, La Y, Bao P, Guo X, Pei J, Yan P, Liang C. Huang C, et al. Cells. 2022 Nov 17;11(22):3654. doi: 10.3390/cells11223654. Cells. 2022. PMID: 36429081 Free PMC article. - Co-expression Network Revealed Roles of RNA m6A Methylation in Human β-Cell of Type 2 Diabetes Mellitus.
Chen C, Xiang Q, Liu W, Liang S, Yang M, Tao J. Chen C, et al. Front Cell Dev Biol. 2021 May 18;9:651142. doi: 10.3389/fcell.2021.651142. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34084770 Free PMC article. - mTORC1-chaperonin CCT signaling regulates m6A RNA methylation to suppress autophagy.
Tang HW, Weng JH, Lee WX, Hu Y, Gu L, Cho S, Lee G, Binari R, Li C, Cheng ME, Kim AR, Xu J, Shen Z, Xu C, Asara JM, Blenis J, Perrimon N. Tang HW, et al. Proc Natl Acad Sci U S A. 2021 Mar 9;118(10):e2021945118. doi: 10.1073/pnas.2021945118. Proc Natl Acad Sci U S A. 2021. PMID: 33649236 Free PMC article. - Reshaping the role of m6A modification in cancer transcriptome: a review.
Yang G, Sun Z, Zhang N. Yang G, et al. Cancer Cell Int. 2020 Jul 29;20:353. doi: 10.1186/s12935-020-01445-y. eCollection 2020. Cancer Cell Int. 2020. PMID: 32760220 Free PMC article. Review.
References
- Beemon K, Keith J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J Mol Biol. 1977;113:165–179. - PubMed
- Bringmann P, Luhrmann R. Antibodies specific for N6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS Lett. 1987;213:309–315. - PubMed
- Brockman JM, Singh P, Liu D, Quinlan S, Salisbury J, Graber JH. PACdb: PolyA Cleavage Site and 3'-UTR Database. Bioinformatics. 2005;21:3691–3693. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 MH095353/MH/NIMH NIH HHS/United States
- T32 CA062948/CA/NCI NIH HHS/United States
- T32CA062948/CA/NCI NIH HHS/United States
- 1F32MH095353-01/MH/NIMH NIH HHS/United States
- 1R01NS076465-01/NS/NINDS NIH HHS/United States
- MH080420/MH/NIMH NIH HHS/United States
- R01 NS076465/NS/NINDS NIH HHS/United States
- R01 MH080420/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources