Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism - PubMed (original) (raw)
Comparative Study
. 2012 Jun 5;109(23):8954-8.
doi: 10.1073/pnas.1200966109. Epub 2012 May 18.
Affiliations
- PMID: 22611190
- PMCID: PMC3384155
- DOI: 10.1073/pnas.1200966109
Comparative Study
Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism
David Veesler et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Phages of the Caudovirales order possess a tail that recognizes the host and ensures genome delivery upon infection. The X-ray structure of the approximately 1.8 MDa host adsorption device (baseplate) from the lactococcal phage TP901-1 shows that the receptor-binding proteins are pointing in the direction of the host, suggesting that this organelle is in a conformation ready for host adhesion. This result is in marked contrast with the lactococcal phage p2 situation, whose baseplate is known to undergo huge conformational changes in the presence of Ca(2+) to reach its active state. In vivo infection experiments confirmed these structural observations by demonstrating that Ca(2+) ions are required for host adhesion among p2-like phages (936-species) but have no influence on TP901-1-like phages (P335-species). These data suggest that these two families rely on diverse adhesion strategies which may lead to different signaling for genome release.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
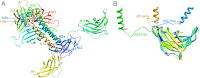
Fig. 1.
Structure of the L. lactis phage TP901-1 baseplate (i.e., host adsorption machinery). (A, Left) Negative-stained electron micrograph of a TP901-1 virion. (Right) Close-up view of the phage baseplate X-ray structure fitted in the adsorption device three-dimensional electron microscopy reconstruction (the region is highlighted by a black square on the micrograph). The baseplate is formed by 18 copies of BppU (red) arranged around a central Dit hexamer (green) and holding eighteen trimeric RBPs (receptor-binding proteins, blue). (B) Top view of the baseplate looking down the phage tail tube axis. (C) Semitilted view of the baseplate rotated by 30° relative to A. (D) The three different types of polypeptides present in the organelle. (Left) A BppU trimer, (Center) the Dit hexamer, and (Right) a RBP trimer. One monomer within each protein homooligomer is depicted in dark gray.
Fig. 2.
Interactions between the different TP901-1 baseplate proteins. (A) The six BppU trimers (red) form a crown around the central Dit hexamer (green). The threefold symmetry axis present within the BppU small helical segments and C-terminal domains (residues 180–299) is evidenced for one BppU trimer. (B) Top view of the baseplate looking down the phage tail tube axis (the RBP trimers have been removed for clarity). Each BppU trimer exhibits an asymmetric organization emphasized through coloring the three nonequivalent BppU chains with a shade of blue, green, and red. The BppU N-terminal domain dodecameric ring surrounding Dit (depicted in light gray) is composed of the yellow (Nt1) and blue (Nt2) domains. (C) Close-up view of the interactions established between BppU and Dit (light gray). The three BppU N-terminal domains are labeled Nt1 to Nt3 with Nt1 and Nt2 forming the dodecameric ring whereas Nt3 interacts with the adjacent BppU trimer. Note the differences in both structure and length observed at the level of the linkers connecting Nt domains with the long helices. The Dit C-terminal domains have been removed for clarity. (D, Left) Each RBP trimer (light gray) is anchored to the baseplate via a loop extending from each BppU C-terminal domain (orange) that penetrates the cup formed at the top of this former protein. (Right) Close-up view of the electrostatic surface potential of the interacting regions from BppU (Upper) and the RBP (Lower) highlighting their high charge and surface complementarity. Multiple sequence alignments of BppU proteins (E) and RBPs (F) from phages TP901-1, P335, ul36, and Tuc2009. Only the region corresponding to the RBP-interacting loop (BppU residues 211–280) is shown. The conserved amino-acids involved in the interactions between the two proteins are indicated by a purple circle for aliphatic/aromatic residues or a blue circle for charged residues.
Fig. 3.
The BppU (ORF48) structure. (A) One BppU trimer is depicted to emphasize on the kink responsible for rephasing of the amino acid residues between the three monomers. Each monomer is colored with a shade of blue, green, or red and the individual domains or segments are annotated. (B) The three BppU N-terminal domains belonging to a single trimer have been superimposed to highlight the difference in both structure and length between their linkers connecting to the long helices.
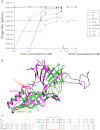
Fig. 4.
Effect of Ca2+ ions on lactococcal phages infectivity in vivo. (A) Phage titre [expressed as plaque-forming units per milliliter (pfu/mL)] for virions of the 936-species (p2, bIL170, and sk1) or P335-species (TP901-1, Tuc2009, P335, and ul36) in the presence of varying concentrations of Ca2+ ions. The phages from the 936 family strictly depend on this divalent cation for infectivity whereas P335-phages are fully infectious in the absence of such ions (with the exception of Tuc2009 that exhibits a partial infectivity in the absence of Ca2+ that could be explained by its different baseplate composition). (B) Superimposition of the Dit N-terminal domains of phages TP901-1 (green) and p2 (pink) showing the absence of the Ca2+ binding loop in this former phage (red arrow) and explaining the difference of phenotype between the two phage species (P335 and 936). (C) Structure-based sequence alignment of the Dit sequences from the 936 and P335 phages used in the in vivo assay based on the structural superimposition in B.
Similar articles
- The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Legrand P, Collins B, Blangy S, Murphy J, Spinelli S, Gutierrez C, Richet N, Kellenberger C, Desmyter A, Mahony J, van Sinderen D, Cambillau C. Legrand P, et al. mBio. 2016 Jan 26;7(1):e01781-15. doi: 10.1128/mBio.01781-15. mBio. 2016. PMID: 26814179 Free PMC article. - Identification of the lower baseplate protein as the antireceptor of the temperate lactococcal bacteriophages TP901-1 and Tuc2009.
Vegge CS, Vogensen FK, Mc Grath S, Neve H, van Sinderen D, Brøndsted L. Vegge CS, et al. J Bacteriol. 2006 Jan;188(1):55-63. doi: 10.1128/JB.188.1.55-63.2006. J Bacteriol. 2006. PMID: 16352821 Free PMC article. - The tal gene of lactococcal bacteriophage TP901-1 is involved in DNA release following host adsorption.
Ruiz-Cruz S, Erazo Garzon A, Cambillau C, Ortiz Charneco G, Lugli GA, Ventura M, Mahony J, van Sinderen D. Ruiz-Cruz S, et al. Appl Environ Microbiol. 2024 Sep 18;90(9):e0069424. doi: 10.1128/aem.00694-24. Epub 2024 Aug 12. Appl Environ Microbiol. 2024. PMID: 39132999 Free PMC article. - Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors.
Goulet A, Spinelli S, Mahony J, Cambillau C. Goulet A, et al. Viruses. 2020 May 6;12(5):512. doi: 10.3390/v12050512. Viruses. 2020. PMID: 32384698 Free PMC article. Review. - Structures and host-adhesion mechanisms of lactococcal siphophages.
Spinelli S, Veesler D, Bebeacua C, Cambillau C. Spinelli S, et al. Front Microbiol. 2014 Jan 16;5:3. doi: 10.3389/fmicb.2014.00003. eCollection 2014. Front Microbiol. 2014. PMID: 24474948 Free PMC article. Review.
Cited by
- Viral infection modulation and neutralization by camelid nanobodies.
Desmyter A, Farenc C, Mahony J, Spinelli S, Bebeacua C, Blangy S, Veesler D, van Sinderen D, Cambillau C. Desmyter A, et al. Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):E1371-9. doi: 10.1073/pnas.1301336110. Epub 2013 Mar 25. Proc Natl Acad Sci U S A. 2013. PMID: 23530214 Free PMC article. - Partial Atomic Model of the Tailed Lactococcal Phage TP901-1 as Predicted by AlphaFold2: Revelations and Limitations.
Mahony J, Goulet A, van Sinderen D, Cambillau C. Mahony J, et al. Viruses. 2023 Dec 15;15(12):2440. doi: 10.3390/v15122440. Viruses. 2023. PMID: 38140681 Free PMC article. - Crystal structure of pb9, the distal tail protein of bacteriophage T5: a conserved structural motif among all siphophages.
Flayhan A, Vellieux FM, Lurz R, Maury O, Contreras-Martel C, Girard E, Boulanger P, Breyton C. Flayhan A, et al. J Virol. 2014 Jan;88(2):820-8. doi: 10.1128/JVI.02135-13. Epub 2013 Oct 23. J Virol. 2014. PMID: 24155371 Free PMC article. - Complete genomes and comparative analyses of Streptomyces phages that influence secondary metabolism and sporulation.
Kronheim S, Solomon E, Ho L, Glossop M, Davidson AR, Maxwell KL. Kronheim S, et al. Sci Rep. 2023 Jun 17;13(1):9820. doi: 10.1038/s41598-023-36938-z. Sci Rep. 2023. PMID: 37330527 Free PMC article. - Identification of a new P335 subgroup through molecular analysis of lactococcal phages Q33 and BM13.
Mahony J, Martel B, Tremblay DM, Neve H, Heller KJ, Moineau S, van Sinderen D. Mahony J, et al. Appl Environ Microbiol. 2013 Jul;79(14):4401-9. doi: 10.1128/AEM.00832-13. Epub 2013 May 10. Appl Environ Microbiol. 2013. PMID: 23666331 Free PMC article.
References
- Kostyuchenko VA, et al. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10:688–693. - PubMed
- Kanamaru S, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–557. - PubMed
- Kostyuchenko VA, et al. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol. 2005;12:810–813. - PubMed
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous