Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing - PubMed (original) (raw)
Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing
Madapura M Pradeepa et al. PLoS Genet. 2012.
Abstract
Increasing evidence suggests that chromatin modifications have important roles in modulating constitutive or alternative splicing. Here we demonstrate that the PWWP domain of the chromatin-associated protein Psip1/Ledgf can specifically recognize tri-methylated H3K36 and that, like this histone modification, the Psip1 short (p52) isoform is enriched at active genes. We show that the p52, but not the long (p75), isoform of Psip1 co-localizes and interacts with Srsf1 and other proteins involved in mRNA processing. The level of H3K36me3 associated Srsf1 is reduced in Psip1 mutant cells and alternative splicing of specific genes is affected. Moreover, we show altered Srsf1 distribution around the alternatively spliced exons of these genes in Psip1 null cells. We propose that Psip1/p52, through its binding to both chromatin and splicing factors, might act to modulate splicing.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
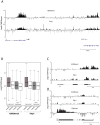
Figure 1. Psip1 PWWP domain binds to H3K36me3.
A) Diagram of p52 and p75 Psip1 isoforms showing the position of the; PWWP domain, AT hook-like domains (hatched box), C-terminal 8 a.a. unique to p52 (black box), and the p75-specific IBD. Vertical arrow indicates the site of gene trap integration in Psip1gt/gt. Horizontal lines indicate the position of epitopes recognized by antibodies A300-847 and A300-848. B) Peptide array containing 384 histone tail modification combinations incubated with GST-Psip1-PWWP and detected with αGST. Spots corresponding to unmodified H3 26–45 peptide (arrow) and H3K36me3 (arrowhead) are indicated. C) Binding specificity (calculated from the intensity of the histone peptide interaction) of Psip1-PWWP (y axis) to the top list of histone modifications arranged according to decreasing specificity (x axis). Data for all the modifications are provided in Table S1. D) Immunoblot of biotinylated H3K36me3 peptide pull-down detecting GST-p52 with αGST antibodies. Corresponding unmodified histone H3 peptide served as control and GST-p52 was loaded as input. E) Immunoblot of A300-847 IPs with antibodies detecting; unmodified H3, H3K36me3, H3K9me2 and H3K4me3. IgG served as control and 5% of NIH3T3 nuclear extract was loaded as input.
Figure 2. Genomic distribution of Psip1/p52 and H3K36me3.
A) Mean log2 ChIP∶input for Psip1/p52 and H3K36me3 in MEFs for an approximately 1.2Mb genomic window from mouse chromosome 5. n = 4 (3 biological and 1 technical replicate). B) Box plots showing the distribution of log2 ChIP∶input for Psip1/p52 and H3K36me3 across exons and introns of expressed or non-expressed genes. Data are deposited in NCBI GEO (Accession no. GSM697402-GSM697411). C, D) Mean log2 ChIP∶input for Psip1/p52 and H3K36me3 in MEFs at (C) c-Myc and (D) Xist loci. H3K4me3 is also shown for XIST. Filled boxes indicate the positions of exons. n = 4 (3 biological and 1 technical replicate) for H3K36me3 and Psip1. NCBI GEO accession number for array platform is GPL13276. n = 2 biological: replicates for H3K4me3.
Figure 3. Immunoprecipitation of Psip1/p52 and p75.
A) Immunoblot of NIH 3T3 nuclear extract with antibodies; A300-848 which recognizes only the p75 isoform of Psip1, and A300-847 which detects both p52 and p75. B) IPs with IgG, A300-847 and A300-848 from NIH 3T3 nuclear extracts, immunoblotted with antibodies recognizing p75 (A300-848) or p52 (A300-847). Input is 5% total extract. C) Immunoblot of A300-847 IPs with αSRSF1. IP was also performed in the presence of RNase A. Input is 10% of total extract and IgG served as a control. D) In vitro pulldown of 293T cell expressed T7-SRSF1 using GST-p52 and Psip1-PWWP and immunoblotted with αT7. Input is 5% of T7-SRSF1 and GST alone is control. E) In vitro pulldown with GST-p52 of; T7-SRSF1 and mutants that mimic its hypo-(RG) and hyper-phosporylation (RD), T7-SRSF3 and GFP-SRSF2. Immunoblotting was with αT7 or αGFP. F) ChIP with αH3K36me3 from wild-type (wt) and Psip1 −/− MEFs immunoblotted with antibodies detecting Srsf1, Srsf2, Srsf3, PTB, Psip1 and H3K36me3. G) In vitro pulldown of HeLa core histones by T7-SRSF1 in the presence or absence of Psip1/p52 and immunoblotted with antibodies detecting pan H3, H3K36me3, H3K9me2 and H3K4me3.
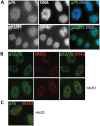
Figure 4. Sub-cellular localization of Psip1/p52 and p75.
A) Immunofluorescence and wide-field epifluorescence microscopy on human cells with; (upper row) p75-specific antibody A300-848, (lower row) A300-847 which can recognize both p52 and p75. DNA was counterstained with DAPI. B) Co-immunofluorescence of Psip1/p52 (green/A300-847) and SRSF2 (red) analyzed by confocal microscopy in untreated (upper row), or actinomycin D (ActD) treated cells. C) Co-immunofluorescence of Psip1/p75 (green/A300-848) and SRSF2 (red) in ActD treated cells and analyzed by confocal microscopy.
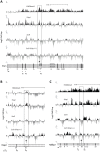
Figure 5. Alternative splicing in Psip1 gt/gt cells.
RT-PCR to detect; exon inclusion (In) or skipping (Skip) of (A); Ptprc, Ppfibp1, Rapgef6, Rasgrp3, Ogfrl, and Sorb2 all of which showed evidence for altered alternative splicing in array analysis, or (B) Csnk1d and Alg9, which were unchanged in the array, in RNAs prepared from wt and Psip1 gt/gt primary MEFs. C) Specific exon-exon junctions (constitutive-constitutive or constitutive-alternative) of Vcan, Tpp2 and Diap2 in wt and Psip1 gt/gt MEFs. D) Specific exon-exon junctions (constitutive-constitutive) of Vcan (5′ exons) and Diap2 (3′exons) and constitutive-constitutive (5′) and constitutive-alternative of Tpp2 alternative exon 24 in wt and Psip1 gt/gt MEFs. E) Specific exon-exon junctions (constitutive-constitutive or constitutive-alternative) of Vcan, Tpp2 and Diap2; exon inclusion (In) or skipping (Skip) of Sorb2 in wt and Psip1 −/− MEFs, and after transfection of p52 or p75 Psip1 into Psip1 −/− MEFs. F) Exon inclusion (In) or skipping (Skip) of; Csnk1d Alg9, and constitutive-constitutive (5′) and constitutive-alternative of Tpp2 alternative exon 24 in RNAs prepared from wt and Psip1 −/− MEFs, and after transfection of p52 or p75 Psip1 into Psip1 −/− MEFs. Sequence and position of primer pairs for each exons are listed in Table S3. Below the gels in panels A to F, the mean ratio of alternative∶constitutive exon (+/− s.e.m.) is shown for three biological replicates. G) Immunoblots of proteins using A300-847 antibodies to detect p75 and p52 in wt and Psip1 −/− MEFs, also Psip1 −/− MEFs transfected with p52 (Psip1 −/− p52Res) and p75 (Psip1 −/− p75Res). Immunoblot with Pcna served as a loading control.
Figure 6. ChIP for H3K36me3, Psip1, and Srsf1 in wt and Psip1 mutant MEFs.
A) Mean log2 ChIP∶input for H3K36me3, Psip1 and Srsf1 in wt MEFs across the Vcan (A), Diap2 (B) and Ppfibp1 (C) loci. Distribution of Srsf1 in chromatin from Psip1 −/− MEFs is also shown. Filled boxes indicate the positions of exons and the arrows indicate the position of alternatively spliced exons whose inclusion into spliced mRNAs is altered in Psip1 gt/gt cells. n = 2 biological replicates that also incorporate a technical (dye-swap) replicate. Array platform number is GPL14175 and the GEO accession numbers for ChIP data are; GSM782590 (Psip1), GSM782591 (H3K36me3), GSM782592 and GSM782593 (Srsf1 in wt), GSM782594 and GSM782595 (Srsf1 in Psip1 −/−).
Similar articles
- Embryonic Lethality Due to Arrested Cardiac Development in Psip1/Hdgfrp2 Double-Deficient Mice.
Wang H, Shun MC, Dickson AK, Engelman AN. Wang H, et al. PLoS One. 2015 Sep 14;10(9):e0137797. doi: 10.1371/journal.pone.0137797. eCollection 2015. PLoS One. 2015. PMID: 26367869 Free PMC article. - H3K36me3 and PSIP1/LEDGF associate with several DNA repair proteins, suggesting their role in efficient DNA repair at actively transcribing loci.
Sundarraj J, Taylor GCA, von Kriegsheim A, Pradeepa MM. Sundarraj J, et al. Wellcome Open Res. 2021 Sep 14;2:83. doi: 10.12688/wellcomeopenres.11589.4. eCollection 2017. Wellcome Open Res. 2021. PMID: 34541330 Free PMC article. - Disruption of Ledgf/Psip1 results in perinatal mortality and homeotic skeletal transformations.
Sutherland HG, Newton K, Brownstein DG, Holmes MC, Kress C, Semple CA, Bickmore WA. Sutherland HG, et al. Mol Cell Biol. 2006 Oct;26(19):7201-10. doi: 10.1128/MCB.00459-06. Mol Cell Biol. 2006. PMID: 16980622 Free PMC article. - Role of LEDGF/p75 (PSIP1) in oncogenesis. Insights in molecular mechanism and therapeutic potential.
Akele M, Iervolino M, Van Belle S, Christ F, Debyser Z. Akele M, et al. Biochim Biophys Acta Rev Cancer. 2025 Feb;1880(1):189248. doi: 10.1016/j.bbcan.2024.189248. Epub 2024 Dec 18. Biochim Biophys Acta Rev Cancer. 2025. PMID: 39701326 Review. - Design principles of interconnections between chromatin and pre-mRNA splicing.
de Almeida SF, Carmo-Fonseca M. de Almeida SF, et al. Trends Biochem Sci. 2012 Jun;37(6):248-53. doi: 10.1016/j.tibs.2012.02.002. Epub 2012 Mar 5. Trends Biochem Sci. 2012. PMID: 22398209 Review.
Cited by
- Chromatin organization at the nuclear pore favours HIV replication.
Lelek M, Casartelli N, Pellin D, Rizzi E, Souque P, Severgnini M, Di Serio C, Fricke T, Diaz-Griffero F, Zimmer C, Charneau P, Di Nunzio F. Lelek M, et al. Nat Commun. 2015 Mar 6;6:6483. doi: 10.1038/ncomms7483. Nat Commun. 2015. PMID: 25744187 Free PMC article. - Gene body methylation in cancer: molecular mechanisms and clinical applications.
Wang Q, Xiong F, Wu G, Liu W, Chen J, Wang B, Chen Y. Wang Q, et al. Clin Epigenetics. 2022 Nov 28;14(1):154. doi: 10.1186/s13148-022-01382-9. Clin Epigenetics. 2022. PMID: 36443876 Free PMC article. Review. - Methylation of histone H3 lysine 36 is a barrier for therapeutic interventions of head and neck squamous cell carcinoma.
Caeiro LD, Nakata Y, Borges RL, Garcia-Martinez L, Bañuelos CP, Stransky S, Chan HL, Brabson J, Domínguez D, Zhang Y, Lewis PW, Aznar-Benitah S, Cimmino L, Bilbao D, Sidoli S, Verdun RE, Morey L. Caeiro LD, et al. bioRxiv [Preprint]. 2023 Nov 6:2023.11.06.565847. doi: 10.1101/2023.11.06.565847. bioRxiv. 2023. PMID: 38076924 Free PMC article. Updated. Preprint. - SETD2: an epigenetic modifier with tumor suppressor functionality.
Li J, Duns G, Westers H, Sijmons R, van den Berg A, Kok K. Li J, et al. Oncotarget. 2016 Aug 2;7(31):50719-50734. doi: 10.18632/oncotarget.9368. Oncotarget. 2016. PMID: 27191891 Free PMC article. Review. - Chromatin's thread to alternative splicing regulation.
Iannone C, Valcárcel J. Iannone C, et al. Chromosoma. 2013 Dec;122(6):465-74. doi: 10.1007/s00412-013-0425-x. Epub 2013 Aug 3. Chromosoma. 2013. PMID: 23912688 Review.
References
- Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. - PubMed
- Cramer P, Caceres JF, Cazalla D, Kadener S, Muro AF, et al. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell. 1999;4:251–258. - PubMed
- Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5:e1000682. doi: 10.1371/journal.pgen.1000682. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 089701/WT_/Wellcome Trust/United Kingdom
- MC_PC_U127527202/MRC_/Medical Research Council/United Kingdom
- MC_U105185858/MRC_/Medical Research Council/United Kingdom
- WT085767/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous