GDP-mannose-4,6-dehydratase is a cytosolic partner of tankyrase 1 that inhibits its poly(ADP-ribose) polymerase activity - PubMed (original) (raw)
GDP-mannose-4,6-dehydratase is a cytosolic partner of tankyrase 1 that inhibits its poly(ADP-ribose) polymerase activity
Kamlesh K Bisht et al. Mol Cell Biol. 2012 Aug.
Abstract
Tankyrase 1 is a poly(ADP-ribose) polymerase (PARP) that participates in a broad range of cellular activities due to interaction with multiple binding partners. Tankyrase 1 recognizes a linear six-amino-acid degenerate motif and, hence, has hundreds of potential target proteins. Binding of partner proteins to tankyrase 1 usually results in their poly(ADP-ribosyl)ation (PARsylation) and can lead to ubiquitylation and proteasomal degradation. However, it is not known how tankyrase 1 PARP activity is regulated. Here we identify GDP-mannose 4,6-dehydratase (GMD) as a binding partner of tankyrase 1. GMD is a cytosolic protein required for the first step of fucose synthesis. We show that GMD is complexed to tankyrase 1 in the cytosol throughout interphase, but its association with tankyrase 1 is reduced upon entry into mitosis, when tankyrase 1 binds to its other partners TRF1 (at telomeres) and NuMA (at spindle poles). In contrast to other binding partners, GMD is not PARsylated by tankyrase 1. Indeed, we show that GMD inhibits tankyrase 1 PARP activity in vitro, dependent on the GMD tankyrase 1 binding motif. In vivo, depletion of GMD led to degradation of tankyrase 1, dependent on the catalytic PARP activity of tankyrase 1. We speculate that association of tankyrase 1 with GMD in the cytosol sequesters tankyrase 1 in an inactive stable form that can be tapped by other target proteins as needed.
Figures
Fig 1
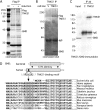
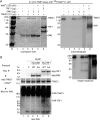
GMD is a novel binding partner of tankyrase 1. (A and B) GMD coimmunoprecipitated with FlagTNKS1 using a (A) one- or (B) two-step protocol. (A) Coomassie-stained gel of proteins immunoprecipitated from an HTC75 cell line (F7) expressing inducible FlagTNKS1 that was grown with (+) or without (−) induction and subject to immunoprecipitation with anti-Flag beads. (B) Coomassie-stained gel of proteins isolated from two rounds of immunoprecipitation from HeLa S3 cell lines stably expressing a vector (V) or FlagTNKS1. Cell lysates were immunoprecipitated with anti-Flag beads, bound proteins were eluted with Flag peptide, and the eluates were subject to immunoprecipitation with anti-TNKS1 762 antibody. (A and B) The band indicated as GMD was excised and its identity was determined by mass spectrometry. (C) Endogenous GMD coimmunoprecipitated with endogenous tankyrase 1. HeLaI.2.11 cell lysates were immunoprecipitated with control or anti-TNKS1 762 antibodies and analyzed by immunoblotting with anti-TNKS1 762 or anti-GMD antibodies. (D) Schematic diagram comparing bacterial and human GMD. Human GMD has an amino-terminal extension that contains the TNKS-binding motif RDSGDG. Below, alignment of the amino termini of GMD from Escherichia coli (NCBI protein accession number ADV17654), Homo sapiens (protein accession number AAH00117), Pan troglodytes (reference sequence number XP_518203), Macaca mulatta (protein accession number AFE65973), Callithrix jacchus (reference sequence number XP_002746325), Crisetulus grieseus (reference sequence number NP_001233625), Rattus norvegicus (protein accession number AAI04709), Mus musculus (protein accession number AAH93502), Bos taurus (protein accession number AAI03031), Xenopus laevis (protein accession number AAI57412), and Danio rerio (protein accession number BAF73663). Identical amino acids are in black.
Fig 2
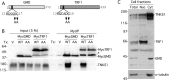
The RGSGDG motif in GMD is required for binding to tankyrase 1. (A) Schematic diagram showing the TNKS binding motif in GMD and TRF1 with the double point mutations indicated. (B) The TNKS binding motif is required for binding of GMD and TRF1. Lysates from HeLaI.2.11 cells transfected with vector (V) or MycGMD (WT or AA) or MycTRF1 (WT or AA) were immunoprecipitated with anti-myc beads and analyzed by immunoblotting with anti-TNKS1 762 or anti-Myc antibodies. *, breakdown product of MycTRF1. (C) Tankyrase 1 binding partners TRF1 and GMD fractionate to distinct subcellular compartments. HeLaI.2.11 cells were fractionated into nuclear and cytosol extracts and analyzed by immunoblotting with anti-TNKS 762, anti-TRF1 415, anti-GMD, and anti-α-tubulin antibodies.
Fig 3
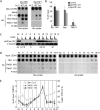
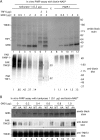
Tankyrase 1 is recruited to telomeres by TRF1 and localizes there in G2/M. (A and B) TRF1 recruits tankyrase 1 to telomeres. Telomeric DNA ChIP analysis of HeLaI.2.11 cells mock transfected (C) or transfected with MycTRF1 (WT or AA) using the indicated beads or antibodies: beads (protein G-Sepharose), Myc beads (Myc-agarose), TRF1 415 (raised against baculovirus-derived full-length TRF1), TNKS1 465 (raised against E. coli-derived tankyrase 1 amino acids 973 to 1149), PI 465 (preimmune serum). Dot blots with the immunoprecipitated DNA were analyzed by Southern blotting with 32P-labeled telomeric or Alu repeat probes. Autoradiographs were cropped from the same experiment. (B) Graphical representation of the percentage of immunoprecipitated telomeric DNA relative to total input DNA derived from three independent experiments; error bars indicate standard deviations. (C to E) Tankyrase localizes to telomeres in G2/M. (C) HeLaI.2.11 cells were synchronized in G1/S by a double thymidine block, released, collected at 2-h intervals, and analyzed by FACS analysis (y axis, cell numbers, 0 to 200; x axis, relative DNA content based on propidium iodide staining, 0 to 600) and by immunoblotting with antibodies against α-tubulin or phospho-histone H3 (Ser10) to mark entry into G2/M. (D) Telomeric DNA ChIP analysis of staged cell cycle extracts using the following antibodies: TRF1 415, TNKS1 465, PI 465, and TNKS1 762 (raised against E. coli-derived tankyrase 1 amino acids 973 to 1149). Dot blots with the immunoprecipitated DNA were analyzed by Southern blotting with 32P-labeled telomeric or Alu repeat probes. Autoradiographs were cropped from the same experiment. (E) Graphical representation of the percentage of immunoprecipitated telomeric DNA relative to total input DNA derived from three independent experiments; error bars indicate standard deviations using the indicated antibodies: anti-TNKS1 465 or 762 antibodies, x axis on the left; anti-TRF1 415 antibody, x axis on the right.
Fig 4
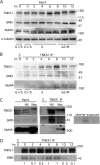
Association of GMD with tankyrase 1 is lost at mitosis. (A and B) Immunoprecipitation analysis across the cell cycle. HeLaI.2.11 cells were synchronized in G1/S by a double thymidine block, released, and collected at 2-h intervals, and whole-cell extracts (Input) generated in TNE buffer were analyzed by immunoblotting with antibodies to TNKS 762, GMD, NuMA, and α-tubulin. *, more slowly migrating form of tankyrase 1 that marks entry into mitosis at the 10- and 12-h time points. (B) Staged extracts generated in TNE buffer were immunoprecipitated with anti-TNKS1 762 and analyzed by immunoblotting with antibodies to TNKS 762, GMD, and NuMA. (C) Immunoprecipitation analysis in cells arrested in S phase versus mitosis. HelaI.2.11 cells were untreated (−) or incubated with hydroxy urea (HU) or nocodazole (Noc) for 20 h. Cell extracts generated in buffer C were analyzed directly (Input) or were immunoprecipitated with control (C) or anti-TNKS1 762 antibody and analyzed by immunoblotting with antibodies to TNKS1 762, GMD, or NuMA. A shorter exposure of the GMD blot is indicated. (D) HeLaI.2.11 cells were synchronized in G1/S by a double thymidine block, released into nocodazole, and collected at 2-h intervals. Staged extracts generated in buffer C were immunoprecipitated with anti-TNKS1 762 and analyzed by immunoblotting with antibodies to TNKS 762 and GMD. GMD levels relative to tankyrase 1 and normalized to the zero time point are indicated below the blot.
Fig 5
GMD is not an acceptor of PARsylation by tankyrase 1. (A) Purified recombinant GMD is not PARsylated by tankyrase 1 in vitro. Recombinant tankyrase 1 was incubated with the substrate 32P-NAD+ in the absence (−) or presence of recombinant GMD or TRF1. Unlabeled NAD+ was added where indicated. The products were fractionated by SDS-PAGE and visualized by Coomassie blue stain (left panel) and autoradiography (right panel; longer exposure, lower right panel). *, breakdown product of MycTRF1. (B) GMD isolated as a tankyrase 1 complex from human cells is not PARsylated by tankyrase 1 in vitro. Extracts from HeLaI.2.11 cells untransfected (−) or transfected with vector (V), MycGMD (WT or AA), or MycTRF1 (WT or AA) were immunoprecipitated with Myc beads. The beads were incubated with recombinant tankyrase 1 and NAD+ substrate, fractionated by SDS-PAGE, and analyzed by immunoblotting with antibodies to Myc (top panel), TNKS1 762 (middle panel), and PAR (lower panel).
Fig 6
GMD inhibits tankyrase 1 PARP activity. (A) GMD inhibits tankyrase 1 but not PARP-1 in vitro. Recombinant tankyrase 1 or PARP-1 was incubated with the substrate biotin-NAD+ in the absence (−) or presence of recombinant GMD and/or TRF1. Proteins were fractionated by SDS-PAGE, transferred to nitrocellulose, and visualized by staining with amido black (top panel) and by immunoblotting with antibiotin antibody (bottom panel). PAR-TNKS1 levels normalized to lane 1 and PAR-PARP-1 levels normalized to lane 8 are indicated below the blot. PAR-TNKS1 and PAR-TRF1 levels in lane 7 are normalized to lane 6 and indicated on the side of the blot. (B) GMD inhibition of tankyrase 1 depends on its TNKS1 binding site. Recombinant tankyrase 1 was incubated with the substrate biotin-NAD+ in the absence or presence of increasing amounts of recombinant GMD (WT or AA). Proteins were fractionated by SDS-PAGE, transferred to nitrocellulose, and visualized by staining with amido black (top panel) and by immunoblotting with anti-biotin (middle panel) or anti-TNKS1 762 antibodies.
Fig 7
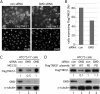
GMD influences tankyrase 1 stability in vivo. (A to C) Induced F7 cells were transfected with control (con) or GMD siRNA for 48 h and analyzed by immunofluorescence (A and B) or immunoblotting (C). (A) Cells were formaldehyde fixed and stained with anti-Flag antibody (top panels) or DAPI (bottom panels). Bar, 100 μm. (B) Graphical representation of the frequency of cells expressing FlagTNKS1 in control versus GMD siRNA cells. Data represent the means ± standard deviations of the results of three independent experiments (n = 300 cells or more each). (C) Depletion of GMD leads to degradation of tankyrase 1 by the proteasome. Cell extracts were analyzed by immunoblotting with anti-Flag, anti-GMD, or anti-α-tubulin antibodies. 12.5 μM MG132 was added 5 h prior to harvest. FlagTNKS1 protein levels relative to α-tubulin and normalized to the siRNA control are indicated below the blot. (D) Loss of FlagTNKS1 depends on its catalytic PARP domain. HTC75 cells were transfected first with control or GMD siRNA for 48 h, followed by transfection with a plasmid expressing FlagTNKS1 wild type (WT) or a PARP-dead allele (PD) for an additional 16 h. Cell extracts were analyzed by immunoblotting with anti-Flag, anti-GMD, or anti-α-tubulin antibodies. FlagTNKS1 protein levels relative to α-tubulin and normalized to the siRNA control are indicated below the blot.
Similar articles
- NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis.
Chang W, Dynek JN, Smith S. Chang W, et al. Biochem J. 2005 Oct 15;391(Pt 2):177-84. doi: 10.1042/BJ20050885. Biochem J. 2005. PMID: 16076287 Free PMC article. - Structural and functional analysis of parameters governing tankyrase-1 interaction with telomeric repeat-binding factor 1 and GDP-mannose 4,6-dehydratase.
Eisemann T, Langelier MF, Pascal JM. Eisemann T, et al. J Biol Chem. 2019 Oct 4;294(40):14574-14590. doi: 10.1074/jbc.RA119.009200. Epub 2019 Aug 2. J Biol Chem. 2019. PMID: 31375564 Free PMC article. - Telomere elongation by a mutant tankyrase 1 without TRF1 poly(ADP-ribosyl)ation.
Muramatsu Y, Tahara H, Ono T, Tsuruo T, Seimiya H. Muramatsu Y, et al. Exp Cell Res. 2008 Mar 10;314(5):1115-24. doi: 10.1016/j.yexcr.2007.12.005. Epub 2007 Dec 14. Exp Cell Res. 2008. PMID: 18221737 - Tankyrase function at telomeres, spindle poles, and beyond.
Hsiao SJ, Smith S. Hsiao SJ, et al. Biochimie. 2008 Jan;90(1):83-92. doi: 10.1016/j.biochi.2007.07.012. Epub 2007 Jul 24. Biochimie. 2008. PMID: 17825467 Review. - TBM Hunter: Identify and Score Canonical, Extended, and Unconventional Tankyrase-Binding Motifs in Any Protein.
Clements CM, Shellman SX, Shellman MH, Shellman YG. Clements CM, et al. Int J Mol Sci. 2023 Nov 30;24(23):16964. doi: 10.3390/ijms242316964. Int J Mol Sci. 2023. PMID: 38069287 Free PMC article. Review.
Cited by
- Tankyrases as modulators of pro-tumoral functions: molecular insights and therapeutic opportunities.
Zamudio-Martinez E, Herrera-Campos AB, Muñoz A, Rodríguez-Vargas JM, Oliver FJ. Zamudio-Martinez E, et al. J Exp Clin Cancer Res. 2021 Apr 28;40(1):144. doi: 10.1186/s13046-021-01950-6. J Exp Clin Cancer Res. 2021. PMID: 33910596 Free PMC article. Review. - TERRA R-loops connect and protect sister telomeres in mitosis.
Sze S, Bhardwaj A, Fnu P, Azarm K, Mund R, Ring K, Smith S. Sze S, et al. Cell Rep. 2023 Oct 31;42(10):113235. doi: 10.1016/j.celrep.2023.113235. Epub 2023 Oct 14. Cell Rep. 2023. PMID: 37843976 Free PMC article. - Cell cycle-regulated ubiquitination of tankyrase 1 by RNF8 and ABRO1/BRCC36 controls the timing of sister telomere resolution.
Tripathi E, Smith S. Tripathi E, et al. EMBO J. 2017 Feb 15;36(4):503-519. doi: 10.15252/embj.201695135. Epub 2016 Dec 19. EMBO J. 2017. PMID: 27993934 Free PMC article. - Persistent telomere cohesion protects aged cells from premature senescence.
Azarm K, Bhardwaj A, Kim E, Smith S. Azarm K, et al. Nat Commun. 2020 Jul 3;11(1):3321. doi: 10.1038/s41467-020-17133-4. Nat Commun. 2020. PMID: 32620872 Free PMC article. - PARP family enzymes: regulation and catalysis of the poly(ADP-ribose) posttranslational modification.
Langelier MF, Eisemann T, Riccio AA, Pascal JM. Langelier MF, et al. Curr Opin Struct Biol. 2018 Dec;53:187-198. doi: 10.1016/j.sbi.2018.11.002. Epub 2018 Nov 24. Curr Opin Struct Biol. 2018. PMID: 30481609 Free PMC article. Review.
References
- Bae J, Donigian JR, Hsueh AJ. 2003. Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J. Biol. Chem. 278:5195–5204 - PubMed
- Becker DJ, Lowe JB. 2003. Fucose: biosynthesis and biological function in mammals. Glycobiology 13:41R–53R - PubMed
- Chang P, Coughlin M, Mitchison TJ. 2005. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 7:1133–1139 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous