Genome characteristics of a novel phage from Bacillus thuringiensis showing high similarity with phage from Bacillus cereus - PubMed (original) (raw)
Comparative Study
Genome characteristics of a novel phage from Bacillus thuringiensis showing high similarity with phage from Bacillus cereus
Yihui Yuan et al. PLoS One. 2012.
Abstract
Bacillus thuringiensis is an important entomopathogenic bacterium belongs to the Bacillus cereus group, which also includes B. anthracis and B. cereus. Several genomes of phages originating from this group had been sequenced, but no genome of Siphoviridae phage from B. thuringiensis has been reported. We recently sequenced and analyzed the genome of a novel phage, BtCS33, from a B. thuringiensis strain, subsp. kurstaki CS33, and compared the gneome of this phage to other phages of the B. cereus group. BtCS33 was the first Siphoviridae phage among the sequenced B. thuringiensis phages. It produced small, turbid plaques on bacterial plates and had a narrow host range. BtCS33 possessed a linear, double-stranded DNA genome of 41,992 bp with 57 putative open reading frames (ORFs). It had a typical genome structure consisting of three modules: the "late" region, the "lysogeny-lysis" region and the "early" region. BtCS33 exhibited high similarity with several phages, B. cereus phage Wβ and some variants of Wβ, in genome organization and the amino acid sequences of structural proteins. There were two ORFs, ORF22 and ORF35, in the genome of BtCS33 that were also found in the genomes of B. cereus phage Wβ and may be involved in regulating sporulation of the host cell. Based on these observations and analysis of phylogenetic trees, we deduced that B. thuringiensis phage BtCS33 and B. cereus phage Wβ may have a common distant ancestor.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Morphology of phage BtCS33 particles under TEM.
The virion was negatively stained with 2% potassium phosphotungstate. The white arrows indicate the putative tail fiber structure.
Figure 2. Genome organization of phage BtCS33.
The schematic represents the whole genome with the ORFs numbered from left to right. Different colors indicate three regions: the “late” region (red color), the “lysogeny-lysis control” region (green color) and the “early” region (blue color). Gray indicates the genes involved in host cell sporulation. Genes with unknown functions are indicated by white. The orientations of the arrows indicate the direction of transcription.
Figure 3. Dot plot alignment of genomes from B. thuringiensis phage BtCS33 and B. cereus phage Wbeta (Wβ).
Arrows indicate the start and the end positions of the most similar fragments on both genomes, corresponding to the dot plot. Both genomes were ranked in the orders of “late” region, the “lysogeney control” region and the “early” region.
Figure 4. Alignment of the proteomes of B. cereus phage Wbeta (Wβ) and B. thuringiensis phage BtCS33.
The putative proteins are numbered and different color arrows show the levels of amino acid identity: green indicates 20%–50%; blue, 50%–80%; and red, 80%–100%.
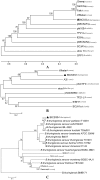
Figure 5. Phylogenetic tree constructed based on the complete geneome and the structural proteins.
(A) Phylogenetic tree constructed from the complete genome sequences of Bacillus spp. phages using a ClustalW alignment and the UPGMA (unweighted pair group method with arithmetic mean) with bootstrap analysis (1,000 replicates). (B) and (C) Phylogenetic trees constructed by using the neighbor-joining method and bootstrap analysis (1,000 replicates) of a Muscle alignment of amino acid sequences of the major capsid proteins and the tail fiber proteins, respectively. The host species is indicated after the name of each phage. ▴ represented the phage isolated in this study. The numbers on the lines indicated the supporting rates. The strains indicated the origin of the tail fiber proteins in (C). The numbers on the lines indicate the branch support.
Similar articles
- Complete genome sequence of the novel phage vB_BthS-HD29phi infecting Bacillus thuringiensis.
Fu Y, Deng S, Liang L, Wu Y, Gao M. Fu Y, et al. Arch Virol. 2019 Dec;164(12):3089-3093. doi: 10.1007/s00705-019-04416-5. Epub 2019 Oct 8. Arch Virol. 2019. PMID: 31595357 - Novel bacteriophages containing a genome of another bacteriophage within their genomes.
Swanson MM, Reavy B, Makarova KS, Cock PJ, Hopkins DW, Torrance L, Koonin EV, Taliansky M. Swanson MM, et al. PLoS One. 2012;7(7):e40683. doi: 10.1371/journal.pone.0040683. Epub 2012 Jul 17. PLoS One. 2012. PMID: 22815791 Free PMC article. - Genome organization of temperate phage 11143 from emetic Bacillus cereus NCTC11143.
Lee YD, Park JH. Lee YD, et al. J Microbiol Biotechnol. 2012 May;22(5):649-53. doi: 10.4014/jmb.1110.10065. J Microbiol Biotechnol. 2012. PMID: 22561859 - Characterization and comparative genomic analysis of bacteriophages infecting members of the Bacillus cereus group.
Lee JH, Shin H, Ryu S. Lee JH, et al. Arch Virol. 2014 May;159(5):871-84. doi: 10.1007/s00705-013-1920-3. Epub 2013 Nov 22. Arch Virol. 2014. PMID: 24264384 Review. - Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: past, present and future.
Gillis A, Mahillon J. Gillis A, et al. Viruses. 2014 Jul 9;6(7):2623-72. doi: 10.3390/v6072623. Viruses. 2014. PMID: 25010767 Free PMC article. Review.
Cited by
- Characterization of bacteriophage vB_KleM_KB2 possessing high control ability to pathogenic Klebsiella pneumoniae.
Peng Q, Ma Z, Han Q, Xiang F, Wang L, Zhang Y, Zhao Y, Li J, Xian Y, Yuan Y. Peng Q, et al. Sci Rep. 2023 Jun 17;13(1):9815. doi: 10.1038/s41598-023-37065-5. Sci Rep. 2023. PMID: 37330608 Free PMC article. - Type 1 Diabetes: an Association Between Autoimmunity, the Dynamics of Gut Amyloid-producing E. coli and Their Phages.
Tetz G, Brown SM, Hao Y, Tetz V. Tetz G, et al. Sci Rep. 2019 Jul 4;9(1):9685. doi: 10.1038/s41598-019-46087-x. Sci Rep. 2019. PMID: 31273267 Free PMC article. - The discovery of phiAGATE, a novel phage infecting Bacillus pumilus, leads to new insights into the phylogeny of the subfamily Spounavirinae.
Barylski J, Nowicki G, Goździcka-Józefiak A. Barylski J, et al. PLoS One. 2014 Jan 23;9(1):e86632. doi: 10.1371/journal.pone.0086632. eCollection 2014. PLoS One. 2014. PMID: 24466180 Free PMC article. - Isolation and characterization of bacteriophages with lytic activity against common bacterial pathogens.
Shende RK, Hirpurkar SD, Sannat C, Rawat N, Pandey V. Shende RK, et al. Vet World. 2017 Aug;10(8):973-978. doi: 10.14202/vetworld.2017.973-978. Epub 2017 Aug 23. Vet World. 2017. PMID: 28919692 Free PMC article. - Isolation of Bacteriophages from Soil Samples in a Poorly Equipped Field Laboratory in Kruger National Park.
Hassim A, Lekota KE. Hassim A, et al. Methods Mol Biol. 2024;2738:91-103. doi: 10.1007/978-1-0716-3549-0_5. Methods Mol Biol. 2024. PMID: 37966593
References
- Jensen GB, Hansen BM, Eilenberg J, Mahillon J. The hidden lifestyles of Bacillus cereus and relatives. Environ Microbiol. 2003;5(8):631–640. - PubMed
- Vilas-Boas GT, Peruca APS, Arantes OMN. Biology and taxonomy of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis. Can J Microbiol. 2007;53(6):673–687. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources