Genetic recombination is directed away from functional genomic elements in mice - PubMed (original) (raw)
Genetic recombination is directed away from functional genomic elements in mice
Kevin Brick et al. Nature. 2012.
Abstract
Genetic recombination occurs during meiosis, the key developmental programme of gametogenesis. Recombination in mammals has been recently linked to the activity of a histone H3 methyltransferase, PR domain containing 9 (PRDM9), the product of the only known speciation-associated gene in mammals. PRDM9 is thought to determine the preferred recombination sites--recombination hotspots--through sequence-specific binding of its highly polymorphic multi-Zn-finger domain. Nevertheless, Prdm9 knockout mice are proficient at initiating recombination. Here we map and analyse the genome-wide distribution of recombination initiation sites in Prdm9 knockout mice and in two mouse strains with different Prdm9 alleles and their F(1) hybrid. We show that PRDM9 determines the positions of practically all hotspots in the mouse genome, with the exception of the pseudo-autosomal region (PAR)--the only area of the genome that undergoes recombination in 100% of cells. Surprisingly, hotspots are still observed in Prdm9 knockout mice, and as in wild type, these hotspots are found at H3 lysine 4 (H3K4) trimethylation marks. However, in the absence of PRDM9, most recombination is initiated at promoters and at other sites of PRDM9-independent H3K4 trimethylation. Such sites are rarely targeted in wild-type mice, indicating an unexpected role of the PRDM9 protein in sequestering the recombination machinery away from gene-promoter regions and other functional genomic elements.
Figures
Figure 1
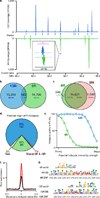
DSB hotspots localize to different loci in 9R and 13R mice. a. DSB hotspots in a representative 0.5 Mb region on chromosome 3. The magenta bar indicates the region shown in the insert. This illustrates two adjacent, yet distinct hotspots in 9R and 13R. Data are smoothed using a 1 Kb sliding window (step 100 bp). Coverage is given as reads per Kb per million (RPKM) b. Overlap between 9R and 13R hotspots. Here and in subsequent panels, only overlaps in the central 400 bp of hotspots are counted. c. Overlap between 9R and C57Bl/6 (B6) hotspots. The excess of B6 hotspots is due to higher ChIP enrichment in this ChIP-Seq sample. B6 hotspots not found in 9R are a weak subset. d. Parental origin of DSB hotspots in the F1 (9R × 13R) mice. e. Stronger 9R and 13R hotspots are active in F1 progeny. The top 14,000 hotspots for each background were binned by strength and the overlap with F1 hotspots assessed for each bin. f. Distribution of hits to the 9R motif (black) and 13R motif (red) around their respective hotspots. Data are plotted in 200 nt steps. g. Alignment of 9R and 13R motifs to the predicted PRDM9 binding sites (see methods). ZNF, Zn finger contact residues; BS, predicted PRDM9 binding site.
Figure 2
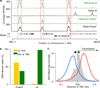
PRDM9 redirects DSBs away from functional genomic elements. a. In _Prdm9_−/− mice DSBs are formed at functional genomic elements marked by H3K4me3 (such as promoters, red boxes). These sites are refractory to DSB formation in wild type mice, where breaks are redirected to PRDM9-dependent H3K4me3 marks (*). b. Most DSBs occur in regions around TSSs in _Prdm9_−/− mice but not in wild type mice. c. The centre of _Prdm9_−/− hotspots at TSS coincides precisely with the H3K4me3 mark on the +1 nucleosome (red circle: −1 nucleosome; white circle: nucleosome free region; green circle: +1 nucleosome). Mean H3K4me3 coverage is grey, mean forward strand ssDNA coverage is blue and reverse strand coverage is red. The intersection point of the forward and reverse strand coverage is the mean hotspot centre (Supplementary Fig. 11).
Figure 3
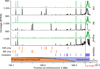
PRDM9-independent hotspots in the PAR and flanking region. Black tracks represent DMC1 tag coverage, green tracks depict H3K4me3 coverage. Orange bars illustrate hotspots unique to 13R or C57Bl/6 (B6) mice. Purple bars illustrate hotspots shared in 13R, C57Bl/6 and _Prdm9_−/− mice.
Figure 4
Proposed role of the PRDM9 protein. Left: In cells containing a functional copy of PRDM9 (Wild type) the DSB formation machinery (scissors) is directed to preferred DSB sites / PRDM9 binding sites. Right: In the absence of PRDM9, the DSB formation machinery opportunistically makes breaks at PRDM9-independent H3K4me3 marks such as those at promoters and enhancers. This results in inefficient DSB repair and meiotic arrest.
Comment in
- Chromosome biology: Pairing up for the genetic exchange.
Stower H. Stower H. Nat Rev Genet. 2012 Jun 18;13(7):449. doi: 10.1038/nrg3265. Nat Rev Genet. 2012. PMID: 22705659 No abstract available.
Similar articles
- The Meiotic Recombination Activator PRDM9 Trimethylates Both H3K36 and H3K4 at Recombination Hotspots In Vivo.
Powers NR, Parvanov ED, Baker CL, Walker M, Petkov PM, Paigen K. Powers NR, et al. PLoS Genet. 2016 Jun 30;12(6):e1006146. doi: 10.1371/journal.pgen.1006146. eCollection 2016 Jun. PLoS Genet. 2016. PMID: 27362481 Free PMC article. - PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice.
Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. Baudat F, et al. Science. 2010 Feb 12;327(5967):836-40. doi: 10.1126/science.1183439. Epub 2009 Dec 31. Science. 2010. PMID: 20044539 Free PMC article. - Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination.
Grey C, Barthès P, Chauveau-Le Friec G, Langa F, Baudat F, de Massy B. Grey C, et al. PLoS Biol. 2011 Oct;9(10):e1001176. doi: 10.1371/journal.pbio.1001176. Epub 2011 Oct 18. PLoS Biol. 2011. PMID: 22028627 Free PMC article. - PRDM9 and Its Role in Genetic Recombination.
Paigen K, Petkov PM. Paigen K, et al. Trends Genet. 2018 Apr;34(4):291-300. doi: 10.1016/j.tig.2017.12.017. Epub 2018 Jan 21. Trends Genet. 2018. PMID: 29366606 Free PMC article. Review. - PRDM9, a driver of the genetic map.
Grey C, Baudat F, de Massy B. Grey C, et al. PLoS Genet. 2018 Aug 30;14(8):e1007479. doi: 10.1371/journal.pgen.1007479. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30161134 Free PMC article. Review.
Cited by
- Crossovers are associated with mutation and biased gene conversion at recombination hotspots.
Arbeithuber B, Betancourt AJ, Ebner T, Tiemann-Boege I. Arbeithuber B, et al. Proc Natl Acad Sci U S A. 2015 Feb 17;112(7):2109-14. doi: 10.1073/pnas.1416622112. Epub 2015 Feb 2. Proc Natl Acad Sci U S A. 2015. PMID: 25646453 Free PMC article. - A Minimal Hybrid Sterility Genome Assembled by Chromosome Swapping Between Mouse Subspecies (Mus musculus).
Fotopulosova V, Tanieli G, Fusek K, Jansa P, Forejt J. Fotopulosova V, et al. Mol Biol Evol. 2024 Oct 4;41(10):msae211. doi: 10.1093/molbev/msae211. Mol Biol Evol. 2024. PMID: 39404090 Free PMC article. - Essential roles of the ANKRD31-REC114 interaction in meiotic recombination and mouse spermatogenesis.
Xu J, Li T, Kim S, Boekhout M, Keeney S. Xu J, et al. Proc Natl Acad Sci U S A. 2023 Nov 21;120(47):e2310951120. doi: 10.1073/pnas.2310951120. Epub 2023 Nov 17. Proc Natl Acad Sci U S A. 2023. PMID: 37976262 Free PMC article. - Zinc finger binding motifs do not explain recombination rate variation within or between species of Drosophila.
Heil CS, Noor MA. Heil CS, et al. PLoS One. 2012;7(9):e45055. doi: 10.1371/journal.pone.0045055. Epub 2012 Sep 18. PLoS One. 2012. PMID: 23028758 Free PMC article. - The pioneering role of PRDM9 indel mutations in tarsier evolution.
Heerschop S, Zischler H, Merker S, Perwitasari-Farajallah D, Driller C. Heerschop S, et al. Sci Rep. 2016 Oct 4;6:34618. doi: 10.1038/srep34618. Sci Rep. 2016. PMID: 27698394 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM084104/GM/NIGMS NIH HHS/United States
- R01 GM084104-01A1/GM/NIGMS NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- 1R01GM084104-01A1/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials