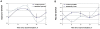Diurnal rhythms in neurexins transcripts and inhibitory/excitatory synapse scaffold proteins in the biological clock - PubMed (original) (raw)
Diurnal rhythms in neurexins transcripts and inhibitory/excitatory synapse scaffold proteins in the biological clock
Mika Shapiro-Reznik et al. PLoS One. 2012.
Abstract
The neurexin genes (NRXN1/2/3) encode two families (α and β) of highly polymorphic presynaptic proteins that are involved in excitatory/inhibitory synaptic balance. Recent studies indicate that neuronal activation and memory formation affect NRXN1/2/3α expression and alternative splicing at splice sites 3 and 4 (SS#3/SS#4). Neurons in the biological clock residing in the suprachiasmatic nuclei of the hypothalamus (SCN) act as self-sustained oscillators, generating rhythms in gene expression and electrical activity, to entrain circadian bodily rhythms to the 24 hours day/night cycles. Cell autonomous oscillations in NRXN1/2/3α expression and SS#3/SS#4 exons splicing and their links to rhythms in excitatory/inhibitory synaptic balance in the circadian clock were explored. NRXN1/2/3α expression and SS#3/SS#4 splicing, levels of neurexin-2α and the synaptic scaffolding proteins PSD-95 and gephyrin (representing excitatory and inhibitory synapses, respectively) were studied in mRNA and protein extracts obtained from SCN of C3H/J mice at different times of the 24 hours day/night cycle. Further studies explored the circadian oscillations in these components and causality relationships in immortalized rat SCN2.2 cells. Diurnal rhythms in mNRXN1α and mNRXN2α transcription, SS#3/SS#4 exon-inclusion and PSD-95 gephyrin and neurexin-2α levels were found in the SCN in vivo. No such rhythms were found with mNRXN3α. SCN2.2 cells also exhibited autonomous circadian rhythms in rNRXN1/2 expression SS#3/SS#4 exon inclusion and PSD-95, gephyrin and neurexin-2α levels. rNRXN3α and rNRXN1/2β were not expressed. Causal relationships were demonstrated, by use of specific siRNAs, between rNRXN2α SS#3 exon included transcripts and gephyrin levels in the SCN2.2 cells. These results show for the first time dynamic, cell autonomous, diurnal rhythms in expression and splicing of NRXN1/2 and subsequent effects on the expression of neurexin-2α and postsynaptic scaffolding proteins in SCN across the 24-h cycle. NRXNs gene transcripts may have a role in coupling the circadian clock to diurnal rhythms in excitatory/inhibitory synaptic balance.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
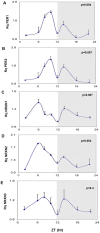
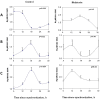
Figure 1. Diurnal changes in mPer1 mPer2 and mNRXN1/2/3α mRNA levels in the mouse SCN.
C3H/J mice kept at 12:12 hours light:dark schedules were sacrificed at different times after light onset (Lights on at time = 0 (ZT0)). mRNA was extracted from the SCN and subjected to real-time PCR. The amount of (A) mPer1, (B) mPer2, (C) Total mNRXN1α, (D) Total mNRXN2α, and (E) Total mNRXN3α transcripts per GAPDH are presented. Values are expressed as Mean +SEM. (N = 3 animals in each time point assessed in quadruplicates). Significance of the rhythm (ANOVA) is presented on each panel. The grey bars indicate the dark phases.
Figure 2. Diurnal changes in mNRXN1/2/3α SS#3/SS#4 exons splicing in the mouse SCN.
Upper panel: Neurexin genes structure and splice sites. Laminin, neurexin, sex-hormone-binding protein (LNS), epidermal growth-factor)-like (EGF), transmembrane (TM) domains, highly glycosylated region (CH), and PDZ-domain-binding site (PDZ BD). The position of each of five alternative splicing sites is indicated (SS#1–SS#5). Exons are identified by numbers; asterisks mark exons subject to alternative splicing. Lower panels: C3H/J mice kept at 12:12 hours light:dark schedules were sacrificed at different times after light onset (Lights on at time = 0 (ZT0)). mRNA was extracted from the SCN and subjected to real-time PCR. The amount of SS#3 exon included transcripts of (A) mNRXN1α, (B) mNRXN2α, (C) mNRXN3α, and of SS#4 exon included transcripts of (D) mNRXN1α, (E) mNRXN2α, (F) mNRXN3α transcripts are expressed per total transcripts of the relevant mNRXN1/2/3α. Values are expressed as Mean +SEM (N = 3 animals in each time point assessed in quadruplicates). Significance of the rhythm (ANOVA) is presented on each panel. The grey bars indicate the dark phases.
Figure 3. Diurnal rhythms in synaptic scaffold proteins and neurexin 2α in the mouse SCN.
C3H/J mice kept at 12:12hours light:dark schedules were sacrificed at different times after light onset (Lights on at time = 0 (ZT0)). Proteins were extracted from the SCN and analyzed by immunoblotting. Representative blots of A) Gephyrin B) PSD-95 and C) neurexin2α and the corresponding GAPDH at various times after lights-on (ZT) are shown in the left panels. Quantifications of PSD-95, gephyrin and neurexin 2α levels relative to GAPDH are presented in the right panel. Values are expressed as Mean +SEM levels of respective protein relative to GAPDH (N = 3 animals in each time point). ZT represents hours from lights-on. The gray shaded areas indicate the dark phase. Significance of the rhythm (ANOVA) is presented on each panel.
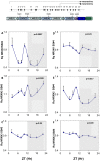
Figure 4. Circadian variations in rPer2, rNRXN1α and rNRXN2α expression in synchronized SCN2.2 cells and the effects of melatonin on them.
Cells (6 plates each) were synchronized and incubated with vehicle (left panels) or 100 nM melatonin (right panels) for 6 hours and collected at different time points during 24 hours (6 plates each). A) rPer2; B) Total rNRXN1α C) Total rNRXN2α qPCR values are normalized vs. rGAPDH. Significance of the rhythms (ANOVA) is denoted on each panel.
Figure 5. Circadian variations in rNRXN1α and rNRXN2α SS#3/SS#4 splicing in SCN2.2 cells and the effects of melatonin on them.
Cells (6 plates each) were synchronized and incubated with vehicle (left panels) or 100 nM melatonin (right panels) for 6 hours and followed up afterwards for 24 hours. The amount of SS#3 exon included transcripts of (A) ) rNRXN1α, (B) rNRXN2α, and of SS#4 exon included transcripts of (C) rNRXN2α, transcripts are expressed per Total transcript of the relevant rNRXN1/2α. Significance of the rhythm (ANOVA) is denoted on each panel.
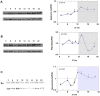
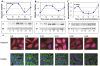
Figure 6. Circadian rhythms in synaptic scaffold protein and neurexin 2α levels and intracellular localization in the SCN2.2 cells.
Cells (6 plates each) were synchronized and collected every 6 hours (6 plates each). Proteins were extracted from the cells and analyzed by SDS PAGE and Western blotting. Protein levels of Gephyrin (A) PSD-95 (B) and neurexin 2α (C) were quantified by immunoblots and normalized to GAPDH levels. Values are Mean +SEM; N = 3 cultures in each time point. Significance of the rhythm (ANOVA) is denoted on each panel. Representative blots of (D) Gephyrin (E) PSD-95 and (F) neurexin2α. The respective GAPDH blots from the same gels (D1, E1 and F1) at various times after synchronization are also depicted. G) SCN2.2 cells were seeded over Poly-L-Lysine coated cover slips, synchronized and fixed by 4% paraformaldehyde after different hours (T = 2–30 h). Cells were immunostained for PSD-95 and Gephyrin, and nuclei were stained with Hoechst staining. An example of membrane staining is noted by a white arrow.
Figure 7. Effects of melatonin on the rhythms in synaptic scaffold protein levels in the SCN2.2 cells.
Cells (6 plates each) were synchronized and incubated with vehicle (circles) or 100 nM melatonin (triangles) for 6 hours and collected every 6 hours (6 plates each). Proteins were extracted from the cells and analyzed by SDS PAGE and Western blotting. Protein levels of Gephyrin (A) and PSD-95 (B) were quantified by immunoblots and normalized to GAPDH levels. Values are Mean +SEM; N = 3 cultures in each time point. Significance of the rhythm (ANOVA) is denoted on each panel.
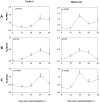
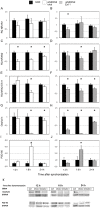
Figure 8. Silencing of NRXN2α and NRXN2α E11 including transcript have differential effects on gephyrin and PSD-95 levels.
Total and E11 including rNRXN2α mRNA levels were suppressed using targeted siRNA sequences (siRNA NRXN2 and siRNA NRXN2 E11 respectively), a scrambled sequence (SCR) was used as a control. Cells were synchronized and then incubated with the siRNAs or SCR for 24 hours and harvested 12,18 or 24 hours afterwards. Mean (+SEM) mRNA levels of (A,B) total rNRXN1α, (C,D) total rNRXN2α, (E, F) E11 including rNRXN2α, and protein levels of (G, H) gephyrin and (I, J) PSD-95 are depicted. Significant differences from control values are marked with asterisks (* p<0.05 t-test against SCR; N = 3). (K) The respective gephyrin, PSD-95 and GAPDH blots are also depicted.
Similar articles
- Oscillating on borrowed time: diffusible signals from immortalized suprachiasmatic nucleus cells regulate circadian rhythmicity in cultured fibroblasts.
Allen G, Rappe J, Earnest DJ, Cassone VM. Allen G, et al. J Neurosci. 2001 Oct 15;21(20):7937-43. doi: 10.1523/JNEUROSCI.21-20-07937.2001. J Neurosci. 2001. PMID: 11588167 Free PMC article. - Dynamic changes in neurexins' alternative splicing: role of Rho-associated protein kinases and relevance to memory formation.
Rozic G, Lupowitz Z, Piontkewitz Y, Zisapel N. Rozic G, et al. PLoS One. 2011 Apr 12;6(4):e18579. doi: 10.1371/journal.pone.0018579. PLoS One. 2011. PMID: 21533271 Free PMC article. - Differential maturation of circadian rhythms in clock gene proteins in the suprachiasmatic nucleus and the pars tuberalis during mouse ontogeny.
Ansari N, Agathagelidis M, Lee C, Korf HW, von Gall C. Ansari N, et al. Eur J Neurosci. 2009 Feb;29(3):477-89. doi: 10.1111/j.1460-9568.2008.06605.x. Eur J Neurosci. 2009. PMID: 19222558 Free PMC article. - GABAergic mechanisms in the suprachiasmatic nucleus that influence circadian rhythm.
Ono D, Honma KI, Honma S. Ono D, et al. J Neurochem. 2021 Apr;157(1):31-41. doi: 10.1111/jnc.15012. Epub 2020 Jul 3. J Neurochem. 2021. PMID: 32198942 Review. - Encoding the ins and outs of circadian pacemaking.
Kuhlman SJ, McMahon DG. Kuhlman SJ, et al. J Biol Rhythms. 2006 Dec;21(6):470-81. doi: 10.1177/0748730406294316. J Biol Rhythms. 2006. PMID: 17107937 Review.
Cited by
- Genome-wide association study of seasonal affective disorder.
Ho KWD, Han S, Nielsen JV, Jancic D, Hing B, Fiedorowicz J, Weissman MM, Levinson DF, Potash JB. Ho KWD, et al. Transl Psychiatry. 2018 Sep 14;8(1):190. doi: 10.1038/s41398-018-0246-z. Transl Psychiatry. 2018. PMID: 30217971 Free PMC article. - Promoter-like sequences regulating transcriptional activity in neurexin and neuroligin genes.
Runkel F, Rohlmann A, Reissner C, Brand SM, Missler M. Runkel F, et al. J Neurochem. 2013 Oct;127(1):36-47. doi: 10.1111/jnc.12372. Epub 2013 Aug 21. J Neurochem. 2013. PMID: 23875667 Free PMC article. - Splice-dependent trans-synaptic PTPδ-IL1RAPL1 interaction regulates synapse formation and non-REM sleep.
Park H, Choi Y, Jung H, Kim S, Lee S, Han H, Kweon H, Kang S, Sim WS, Koopmans F, Yang E, Kim H, Smit AB, Bae YC, Kim E. Park H, et al. EMBO J. 2020 Jun 2;39(11):e104150. doi: 10.15252/embj.2019104150. Epub 2020 Apr 29. EMBO J. 2020. PMID: 32347567 Free PMC article. - Orbitofrontal intronic circular RNA from Nrxn3 mediates reward learning and motivation for reward.
Dabrowski KR, Floris G, Gillespie A, Daws SE. Dabrowski KR, et al. Prog Neurobiol. 2024 Jan;232:102546. doi: 10.1016/j.pneurobio.2023.102546. Epub 2023 Nov 29. Prog Neurobiol. 2024. PMID: 38036039 Free PMC article. - External and circadian inputs modulate synaptic protein expression in the visual system of Drosophila melanogaster.
Krzeptowski W, Górska-Andrzejak J, Kijak E, Görlich A, Guzik E, Moore G, Pyza EM. Krzeptowski W, et al. Front Physiol. 2014 Apr 3;5:102. doi: 10.3389/fphys.2014.00102. eCollection 2014. Front Physiol. 2014. PMID: 24772085 Free PMC article.
References
- Ushkaryov YA, Petrenko AG, Geppert M, Sudhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. - PubMed
- Ushkaryov YA, Hata Y, Ichtchenko K, Moomaw C, Afendis S, et al. Conserved domain structure of beta-neurexins. Unusual cleaved signal sequences in receptor-like neuronal cell-surface proteins. J Biol Chem. 1994;269:11987–11992. - PubMed
- Missler M, Sudhof TC. Neurexins: three genes and 1001 products. Trends Genet. 1998;14:20–26. - PubMed
- Ullrich B, Ushkaryov YA, Sudhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases