Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts - PubMed (original) (raw)
Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts
Catherine Lozupone et al. Genome Res. 2012 Oct.
Abstract
We lack a deep understanding of genetic and metabolic attributes specializing in microbial consortia for initial and subsequent waves of colonization of our body habitats. Here we show that phylogenetically interspersed bacteria in Clostridium cluster XIVa, an abundant group of bacteria in the adult human gut also known as the Clostridium coccoides or Eubacterium rectale group, contains species that have evolved distribution patterns consistent with either early successional or stable gut communities. The species that specialize to the infant gut are more likely to associate with systemic infections and can reach high abundances in individuals with Inflammatory Bowel Disease (IBD), indicating that a subset of the microbiota that have adapted to pioneer/opportunistic lifestyles may do well in both early development and with disease. We identified genes likely selected during adaptation to pioneer/opportunistic lifestyles as those for which early succession association and not phylogenetic relationships explain genomic abundance. These genes reveal potential mechanisms by which opportunistic gut bacteria tolerate osmotic and oxidative stress and potentially important aspects of their metabolism. These genes may not only be biomarkers of properties associated with adaptation to early succession and disturbance, but also leads for developing therapies aimed at promoting reestablishment of stable gut communities following physiologic or pathologic disturbances.
Figures
Figure 1.
Co-occurrence network of human gut bacteria, based on a relative abundance matrix previously reported in Qin et al. (2010). The nodes represent species whose genomes have been sequenced. The size of the nodes indicates the average relative abundance across the 124 individuals in the MetaHIT cohort, and the color of the node reflects taxonomic information. Species with significantly positive co-occurrence for any of six measures used (Pearson, Spearman, Kendall, Bray-Curtis, Euclidean, and mutual information) are joined with an edge. Co-occurrence modules are defined as a set/collection of species that are connected among each other (directly or via several steps), but not to any other species in the network, and are labeled with M_x_. Species' full names are shown in Supplemental Table S1, and phylogenetic relationships among them as determined with 16S rRNA are shown in Supplemental Fig. S1. For further discussion of this network, see the Supplemental Material.
Figure 2.
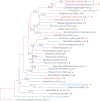
Phylogenetic relationships between species in Clostridium cluster XIVa. Bootstrap support (based on 1000 replicates) is indicated on the branches of the 16S rRNA NJ tree when >40% (except for Ruminococcus sp. SR1/5, as this was added by parsimony insertion after the initial tree creation, since only a short region of 16S rRNA sequence was available; see Methods). The species names are colored according to their module in the co-occurrence network: M1 (blue), M2 (turquoise), M3 (red), and M4 (green). Species that were evaluated in the network analysis but showed no significant co-occurrence are in black text. Species that were not evaluated for co-occurrence are colored purple. These were added to further support that the species in this group that can cause disease form a clade with expanded genome size. The branches are colored by genome size rank, with the red branches representing the largest genomes and the blue branches the smallest. The genome size in Mb is listed after the species name. Species that have been recovered from clinical samples (e.g., bacteremia) are marked with a red circle, and those reported to be in increased abundance with IBD are marked with a blue circle. Species that contain the genomic machinery for a flagellum are marked with a blue curved line. Note that the network diagram only shows species with significant co-occurrence with at least one other species; thus 36.7% (11/30) of evaluated species in Clostridium cluster XIVa, are not shown. Details about the evaluated species are provided in Table 1.
Figure 3.
Relative abundance of species in M1 and M3 in various gut samples. Sample categories are detailed in Supplemental Table S2. (A) Average relative abundance across samples from individuals with and without gastrointestinal disease. (Blue circles) M1 species; (red squares) M3 species. The SEM for each treatment is plotted. For healthy, estimated using data from (1) stool samples from obese and lean twins and their mothers (Turnbaugh et al. 2009a) (Turnbaugh Feces), (2) samples from three healthy adults from six mucosal sites along the length of the colon (Eckburg et al. 2005) (Eckburg Mucosal), (3) fecal samples from the same three individuals as in Eckburg Mucosal (Eckburg Feces). For diseased, averaged relative abundance across (1) colon (Frank Colon), (2) small intestine (Frank SI), and (3) MLNs (Frank MLN) from individuals with gastrointestinal disease including Crohn's disease, ulcerative colitis, and colon cancer from Frank et al. (2007) (B) Results from humanized gnotobiotic mice (Turnbaugh et al. 2009b). Fecal samples from the healthy human donor (donor, [1]) and from the recipient gnotobiotic mice ([2] stomach, [3] small intestine [SI], [4] cecum, [5] colon, and [6] feces). The points representing R. torques, which was an outlier in this analysis, are marked with a green circle. Error bars represent the median and interquartile range. (C) Age trends in M1 and M3 species in a single infant using data from Koenig et al. ( 2011). The _x_-axis in each plot is the age in days and the _y_-axis is the relative abundance in a single sample. The species in M1 have series colored in blue, M3 red, and those with no detected co-occurring microbes are in black. The relative abundance of each OTU in the mother is plotted at day 950 in light green. The period before the introduction of solid food is shaded in blue and between then and the switch from breast milk to formula is shaded in yellow. Ruminococcus sp. SR 1/5 was not evaluated because sequence information for the V2 region of its 16S rRNA is incomplete. Coprococcus comes is not shown because it was absent across the infant timeseries and in the mother.
Figure 4.
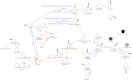
Metabolic network. Generated based on the combined metabolic network of M1 and M3 genomes (see Methods). The nodes (circles) are compounds and the edges are reactions. Edges colored blue have more copies or are more likely present in M3 genomes compared with M1. Red edges are reactions known to occur, but no described enzymes that perform them are in any of these genomes. The node color indicates the fraction of M3 genomes in which that compound is found with white being 100% and black being 0%. The thick green edges indicate that branches of the network emanating from that node were eliminated from the figure.
Similar articles
- Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla.
Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. Mahowald MA, et al. Proc Natl Acad Sci U S A. 2009 Apr 7;106(14):5859-64. doi: 10.1073/pnas.0901529106. Epub 2009 Mar 24. Proc Natl Acad Sci U S A. 2009. PMID: 19321416 Free PMC article. - PCR DGGE and RT-PCR DGGE show diversity and short-term temporal stability in the Clostridium coccoides-Eubacterium rectale group in the human intestinal microbiota.
Maukonen J, Mättö J, Satokari R, Söderlund H, Mattila-Sandholm T, Saarela M. Maukonen J, et al. FEMS Microbiol Ecol. 2006 Dec;58(3):517-28. doi: 10.1111/j.1574-6941.2006.00179.x. FEMS Microbiol Ecol. 2006. PMID: 17117993 - Intestinal colonization: how key microbial players become established in this dynamic process: microbial metabolic activities and the interplay between the host and microbes.
El Aidy S, Van den Abbeele P, Van de Wiele T, Louis P, Kleerebezem M. El Aidy S, et al. Bioessays. 2013 Oct;35(10):913-23. doi: 10.1002/bies.201300073. Epub 2013 Aug 15. Bioessays. 2013. PMID: 23946088 Review. - Links between gut microbiome composition and fatty liver disease in a large population sample.
Ruuskanen MO, Åberg F, Männistö V, Havulinna AS, Méric G, Liu Y, Loomba R, Vázquez-Baeza Y, Tripathi A, Valsta LM, Inouye M, Jousilahti P, Salomaa V, Jain M, Knight R, Lahti L, Niiranen TJ. Ruuskanen MO, et al. Gut Microbes. 2021 Jan-Dec;13(1):1-22. doi: 10.1080/19490976.2021.1888673. Gut Microbes. 2021. PMID: 33651661 Free PMC article. - The first thousand days - intestinal microbiology of early life: establishing a symbiosis.
Wopereis H, Oozeer R, Knipping K, Belzer C, Knol J. Wopereis H, et al. Pediatr Allergy Immunol. 2014 Aug;25(5):428-38. doi: 10.1111/pai.12232. Epub 2014 Jun 5. Pediatr Allergy Immunol. 2014. PMID: 24899389 Review.
Cited by
- Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota.
Smith CC, Snowberg LK, Gregory Caporaso J, Knight R, Bolnick DI. Smith CC, et al. ISME J. 2015 Nov;9(11):2515-26. doi: 10.1038/ismej.2015.64. Epub 2015 Apr 24. ISME J. 2015. PMID: 25909977 Free PMC article. - In Vitro Prebiotic and Anti-Colon Cancer Activities of Agar-Derived Sugars from Red Seaweeds.
Yun EJ, Yu S, Kim YA, Liu JJ, Kang NJ, Jin YS, Kim KH. Yun EJ, et al. Mar Drugs. 2021 Apr 12;19(4):213. doi: 10.3390/md19040213. Mar Drugs. 2021. PMID: 33921308 Free PMC article. - Bacterial flagellin is a dominant, stable innate immune activator in the gastrointestinal contents of mice and rats.
Vijay-Kumar M, Bovilla VR, Yeoh BS, Golonka RM, Saha P, Joe B, Gewirtz AT. Vijay-Kumar M, et al. Gut Microbes. 2023 Jan-Dec;15(1):2185031. doi: 10.1080/19490976.2023.2185031. Gut Microbes. 2023. PMID: 36880647 Free PMC article. - A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease.
Tong M, Li X, Wegener Parfrey L, Roth B, Ippoliti A, Wei B, Borneman J, McGovern DP, Frank DN, Li E, Horvath S, Knight R, Braun J. Tong M, et al. PLoS One. 2013 Nov 19;8(11):e80702. doi: 10.1371/journal.pone.0080702. eCollection 2013. PLoS One. 2013. PMID: 24260458 Free PMC article. - Functional Profiling of Unfamiliar Microbial Communities Using a Validated De Novo Assembly Metatranscriptome Pipeline.
Davids M, Hugenholtz F, Martins dos Santos V, Smidt H, Kleerebezem M, Schaap PJ. Davids M, et al. PLoS One. 2016 Jan 12;11(1):e0146423. doi: 10.1371/journal.pone.0146423. eCollection 2016. PLoS One. 2016. PMID: 26756338 Free PMC article.
References
- Bartlett JG 2002. Clinical practice. Antibiotic-associated diarrhea. N Engl J Med 346: 334–339 - PubMed
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300
Publication types
MeSH terms
Grants and funding
- P01 DK078669/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- DK30292/DK/NIDDK NIH HHS/United States
- R01 HG004872/HG/NHGRI NIH HHS/United States
- DK70977/DK/NIDDK NIH HHS/United States
- T32 GM142607/GM/NIGMS NIH HHS/United States
- HG4866/HG/NHGRI NIH HHS/United States
- DK78669/DK/NIDDK NIH HHS/United States
- R01 DK030292/DK/NIDDK NIH HHS/United States
- HG4872/HG/NHGRI NIH HHS/United States
- R01 DK070977/DK/NIDDK NIH HHS/United States
- T32 GM008759/GM/NIGMS NIH HHS/United States
- U01 HG004866/HG/NHGRI NIH HHS/United States
- K01 DK090285/DK/NIDDK NIH HHS/United States
- K01DK090285/DK/NIDDK NIH HHS/United States
- R37 DK030292/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources