Y chromosome mediates ribosomal DNA silencing and modulates the chromatin state in Drosophila - PubMed (original) (raw)
Y chromosome mediates ribosomal DNA silencing and modulates the chromatin state in Drosophila
Jun Zhou et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Although the Drosophila Y chromosome is degenerated, heterochromatic, and contains few genes, increasing evidence suggests that it plays an important role in regulating the expression of numerous autosomal and X-linked genes. Here we use 15 Y chromosomes originating from a single founder 550 generations ago to study the role of the Y chromosome in regulating rRNA gene transcription, position-effect variegation (PEV), and the link among rDNA copy number, global gene expression, and chromatin regulation. Based on patterns of rRNA gene transcription indicated by transcription of the retrotransposon R2 that specifically inserts into the 28S rRNA gene, we show that X-linked rDNA is silenced in males. The silencing of X-linked rDNA expression by the Y chromosome is consistent across populations and independent of genetic background. These Y chromosomes also vary more than threefold in rDNA locus size and cause dramatically different levels of PEV suppression. The degree of suppression is negatively associated with the number and fraction of rDNA units without transposon insertions, but not with total rDNA locus size. Gene expression profiling revealed hundreds of differentially expressed genes among these Y chromosome introgression lines, as well as a divergent global gene expression pattern between the low-PEV and high-PEV flies. Our findings suggest that the Y chromosome is involved in diverse phenomena related to transcriptional regulation including X-linked rDNA silencing and suppression of PEV phenotype. These results further expand our understanding of the role of the Y chromosome in modulating global gene expression, and suggest a link with modifications of the chromatin state.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
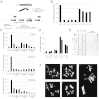
Multiple lines of evidence suggest that the X-linked rDNA is silenced in males. (A) Schematic of the X and Y chromosomes in D. melanogaster and the ribosomal loci containing the retrotransposable element R2. At bottom is a magnified rDNA locus containing R2 elements that insert into specific sites in the 28S rRNA genes. R2 is cotranscribed with the rRNA units; thus, R2 transcription can be used to represent rRNA gene transcription. (B) High levels of expression for the H23 X rDNA locus can be repressed by a low-expressing rDNA loci on another X; however, high levels of expression from the H15 Y rDNA locus cannot be shut off by an X rDNA locus. (C) Nine wild Y chromosomes from three different geographic populations demonstrated preferential Y-linked rDNA expression, also suggesting X-linked rDNA silencing in the males of those crosses. The rDNA locus in w1118 had low transcriptional levels, whereas H23 had high expression levels. (D) Four Y chromosomes with different levels of rDNA and R2 expression were introgressed into the w1118 background. The bar graph shows similar levels of the Y-linked rDNA and R2 expression in the introgressed background compared with their original levels. (E) RT-PCR results demonstrate the expression of specific 5′ truncated R2 elements in males and females. All arrows except the bottom one indicate silencing transcription of specific X-linked R2 copies in males. The bottom arrow shows a transcribed specific R2 copy on the Y chromosome. (F) Premetaphase chromosomes from adult brain tissue stained with DAPI. Cytological analyses found that only the Y chromosome formed a secondary constriction site, indicative of active transcription of the rDNA locus in the previous interphase in males. The Y chromosomes of H8 (a and c), H15 (d and e), and H23 (b) all demonstrated secondary constriction at the rDNA loci, whereas the X chromosome loci did not. Arrows indicate the locations of the rDNA loci on the X and Y chromosomes.
Fig. 2.
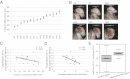
Y-linked rDNA variation correlates with PEV phenotype. (A) The Y-linked rDNA varies more than threefold in size. (B) Representative images showing the variation in levels of eye pigmentation among the Harwich Y introgression lines crossed to the w[m4h] PEV background. (C) The number of transposon uninserted rDNA units is negatively correlated with the PEV phenotype (expression of the w gene measured by eye pigment absorbance at 480 nm). (D) The uninserted fraction of the rDNA locus is negatively correlated with the PEV. (E) Boxplot showing distribution the of log2(low PEV expression) − log2(high PEV expression) for genes encoding ribosomal proteins (based on the Ribosomal Protein Gene database, accessed at
http://ribosome.med.miyazaki-u.ac.jp/
), compared with the distribution of the same fold change for genes not encoding ribosomal proteins. Expression of ribosomal genes is significantly higher in the low-PEV lines (P < 2.2e-16, Mann-Whitney U test).
Fig. 3.
Global gene expression divergence in low-PEV and high-PEV lines. (A) Volcano plot showing more genes up-regulated in red-eyed, low-PEV flies (H15 and H23) compared with white-eyed, high-PEV flies (H5 and H7) than genes down-regulated in that comparison. Black dots indicate differentially expressed genes, whereas gray dots indicate genes that are not differentially expressed. The dots with X-axis values >0 indicate genes that are up-regulated in low-PEV flies; those with values <0 indicate down-regulated genes in low-PEV flies. The GEL50 value as a proxy of power, measuring the gene expression level at which there is a 50% chance of detection of statistical significance, is 1.85-fold in this study (37). (B) The expression data for the two low-PEV and two high-PEV lines were hierarchically clustered using the R package pvclust. A dendrogram shows that H15 and H23 are clustered and H5 and H7 are clustered for global gene expression. (C) To estimate spatial clustering along the genome, the number of significantly up-regulated or down-regulated genes was calculated separately in each 1-MB sliding window across the genome, with a step size of 500 kb. To estimate significance, a number of genes equal to the actual number of significant genes were randomly sampled 10,000 times to get a null expectation for each window. Windows marked with a diamond are significantly elevated at a nominal P < 0.01.
Similar articles
- Ribosomal DNA deletions modulate genome-wide gene expression: "rDNA-sensitive" genes and natural variation.
Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. Paredes S, et al. PLoS Genet. 2011 Apr;7(4):e1001376. doi: 10.1371/journal.pgen.1001376. Epub 2011 Apr 21. PLoS Genet. 2011. PMID: 21533076 Free PMC article. - Roles of rDNA spacer and transcription unit-sequences in X-Y meiotic chromosome pairing in Drosophila melanogaster males.
Ren X, Eisenhour L, Hong C, Lee Y, McKee BD. Ren X, et al. Chromosoma. 1997 Jun;106(1):29-36. doi: 10.1007/s004120050221. Chromosoma. 1997. PMID: 9169584 - An unusual Y chromosome of Drosophila simulans carrying amplified rDNA spacer without rRNA genes.
Lohe AR, Roberts PA. Lohe AR, et al. Genetics. 1990 Jun;125(2):399-406. doi: 10.1093/genetics/125.2.399. Genetics. 1990. PMID: 2379820 Free PMC article. - Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila.
Girton JR, Johansen KM. Girton JR, et al. Adv Genet. 2008;61:1-43. doi: 10.1016/S0065-2660(07)00001-6. Adv Genet. 2008. PMID: 18282501 Review. - The license to pair: identification of meiotic pairing sites in Drosophila.
McKee BD. McKee BD. Chromosoma. 1996 Sep;105(3):135-41. doi: 10.1007/BF02509494. Chromosoma. 1996. PMID: 8781181 Review.
Cited by
- Meta-analysis reveals that genes regulated by the Y chromosome in Drosophila melanogaster are preferentially localized to repressive chromatin.
Sackton TB, Hartl DL. Sackton TB, et al. Genome Biol Evol. 2013;5(1):255-66. doi: 10.1093/gbe/evt005. Genome Biol Evol. 2013. PMID: 23315381 Free PMC article. - Transposable element accumulation drives size differences among polymorphic Y Chromosomes in Drosophila.
Nguyen AH, Wang W, Chong E, Chatla K, Bachtrog D. Nguyen AH, et al. Genome Res. 2022 Jun;32(6):1074-1088. doi: 10.1101/gr.275996.121. Epub 2022 May 2. Genome Res. 2022. PMID: 35501131 Free PMC article. - Integration, Regulation, and Long-Term Stability of R2 Retrotransposons.
Eickbush TH, Eickbush DG. Eickbush TH, et al. Microbiol Spectr. 2015 Apr;3(2):MDNA3-0011-2014. doi: 10.1128/microbiolspec.MDNA3-0011-2014. Microbiol Spectr. 2015. PMID: 26104703 Free PMC article. Review. - What do you mean, "epigenetic"?
Deans C, Maggert KA. Deans C, et al. Genetics. 2015 Apr;199(4):887-96. doi: 10.1534/genetics.114.173492. Genetics. 2015. PMID: 25855649 Free PMC article. - The Y chromosome as a regulatory element shaping immune cell transcriptomes and susceptibility to autoimmune disease.
Case LK, Wall EH, Dragon JA, Saligrama N, Krementsov DN, Moussawi M, Zachary JF, Huber SA, Blankenhorn EP, Teuscher C. Case LK, et al. Genome Res. 2013 Sep;23(9):1474-85. doi: 10.1101/gr.156703.113. Epub 2013 Jun 25. Genome Res. 2013. PMID: 23800453 Free PMC article.
References
- Bull JJ. Evolution of Sex-Determining Mechanisms. London: Benjamin Cummings; 1983.
- Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases