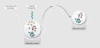Microbiota-targeted therapies: an ecological perspective - PubMed (original) (raw)
Review
Microbiota-targeted therapies: an ecological perspective
Katherine P Lemon et al. Sci Transl Med. 2012.
Abstract
The connection between disease and the disruption of homeostatic interactions between the host and its microbiota is now well established. Drug developers and clinicians are starting to rely more heavily on therapies that directly target the microbiota and on the ecology of the microbiota to understand the outcomes of these treatments. The effects of those microbiota-targeted therapies that alter community composition range in scale from eliminating individual strains of a single species (for example, with antibacterial conjugate vaccines) to replacing the entire community with a new intact microbiota (for example, by fecal transplantation). Secondary infections linked to antibiotic use provide a cautionary tale of the unintended consequences of perturbing a microbial species network and highlight the need for new narrow-spectrum antibiotics with rapid companion diagnostics. Insights into microbial ecology will also benefit the development of probiotics, whose therapeutic prospects will depend on rigorous clinical testing. Future probiotics may take the form of a consortium of long-term community residents: "a fecal transplant in a capsule." The efficacy of microbiota-targeted therapies will need to be assessed using new diagnostic tools that measure community function rather than composition, including the temporal response of a microbial community to a defined perturbation such as an antibiotic or probiotic.
Figures
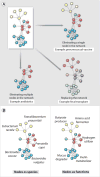
Fig. 1
Microbial communities as networks. (A) Shown are three types of perturbation to a network of microbial species such as that found in the microbiota at various body sites. The microbiota can be perturbed by excision of a single species (node) by a vaccine or a species-specific antibiotic, by elimination of multiple nodes or a subnetwork by an antibiotic, or by replacement of a whole network using microbiota transplantation. (B) Two ways of modeling a microbial community as a network. (Left) Nodes as species, and edges as interactions among species. Species networks can be constructed directly from metagenomic sequence data, but they lack functional information. (Right) Nodes as functions, and edges as interactions among functions. Function networks can generate hypotheses about the mechanism of microbiota-host interactions, but they require mapping genes to functions or a panel of direct functional measurements.
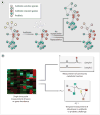
Fig. 2
New opportunities in treatment and diagnostics. (A) Antibiotics save countless lives, but when they kill mutualistic (that is, helpful) microbiota that normally check the growth of pathogens, a secondary infection can ensue. Repopulating antibiotic-treated patients with probiotics is a promising strategy to prevent secondary infections. (B) Given that the human microbiota has many normal taxonomic compositions, it might be easier to develop markers of a normal or healthy community in terms of functional attributes like resistance and resilience. A single time-point measurement of taxon and gene abundance (left) is limited in its ability to provide functional information. Diagnostics based on direct measurements of metabolites (right, top) and temporal measurements of robustness to antibiotic or probiotic challenge (right, bottom) would enable community function to be assessed directly.
Fig. 3
Microbiota-targeted therapy can shift a community to a healthier stable state. A microbial community’s response to a microbiota-targeted therapy can be illustrated with a stability landscape diagram (
). The ball containing the network represents the microbial community, and the shift in its horizontal position within the landscape represents movement between alternative stable states. The depth of a basin indicates the probability that the community will stay in that specific state in response to perturbation and, therefore, reflects the degree of perturbation needed to shift the community to an alternative stable state, for example, from state 1 to state 2. In this illustration, a therapeutic perturbation that removes some nodes (yellow and green) from the community network is sufficient to shift the community to an alternative, and in this case healthier, stable state (state 1).
Similar articles
- Effects of probiotics and antibiotics on the intestinal homeostasis in a computer controlled model of the large intestine.
Rehman A, Heinsen FA, Koenen ME, Venema K, Knecht H, Hellmig S, Schreiber S, Ott SJ. Rehman A, et al. BMC Microbiol. 2012 Mar 27;12:47. doi: 10.1186/1471-2180-12-47. BMC Microbiol. 2012. PMID: 22452835 Free PMC article. - Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken.
Gao P, Ma C, Sun Z, Wang L, Huang S, Su X, Xu J, Zhang H. Gao P, et al. Microbiome. 2017 Aug 3;5(1):91. doi: 10.1186/s40168-017-0315-1. Microbiome. 2017. PMID: 28768551 Free PMC article. - [The microbiota and infectious diarrhea].
Surawicz CM. Surawicz CM. Gastroenterol Clin Biol. 2010 Sep;34 Suppl 1:S29-36. doi: 10.1016/S0399-8320(10)70018-X. Gastroenterol Clin Biol. 2010. PMID: 20889002 French. - Probiotics and antibiotics in IBD.
Sokol H. Sokol H. Dig Dis. 2014;32 Suppl 1:10-7. doi: 10.1159/000367820. Epub 2014 Dec 17. Dig Dis. 2014. PMID: 25531348 Review. - Irritable bowel syndrome: the role of the intestinal microbiota, pathogenesis and therapeutic targets.
Dahlqvist G, Piessevaux H. Dahlqvist G, et al. Acta Gastroenterol Belg. 2011 Sep;74(3):375-80. Acta Gastroenterol Belg. 2011. PMID: 22103040 Review.
Cited by
- Considering humans as habitat reveals evidence of successional disease ecology among human pathogens.
Fefferman NH, Price CA, Stringham OC. Fefferman NH, et al. PLoS Biol. 2022 Sep 12;20(9):e3001770. doi: 10.1371/journal.pbio.3001770. eCollection 2022 Sep. PLoS Biol. 2022. PMID: 36094962 Free PMC article. - Altered Gut Microbiota and Shift in Bacteroidetes between Young Obese and Normal-Weight Korean Children: A Cross-Sectional Observational Study.
Shin S, Cho KY. Shin S, et al. Biomed Res Int. 2020 Aug 18;2020:6587136. doi: 10.1155/2020/6587136. eCollection 2020. Biomed Res Int. 2020. PMID: 32908903 Free PMC article. - Cell-based therapeutics: the next pillar of medicine.
Fischbach MA, Bluestone JA, Lim WA. Fischbach MA, et al. Sci Transl Med. 2013 Apr 3;5(179):179ps7. doi: 10.1126/scitranslmed.3005568. Sci Transl Med. 2013. PMID: 23552369 Free PMC article. - Lactobacillus gasseri LA39 Activates the Oxidative Phosphorylation Pathway in Porcine Intestinal Epithelial Cells.
Hu J, Ma L, Zheng W, Nie Y, Yan X. Hu J, et al. Front Microbiol. 2018 Dec 11;9:3025. doi: 10.3389/fmicb.2018.03025. eCollection 2018. Front Microbiol. 2018. PMID: 30619122 Free PMC article. - Analysis of 16S rRNA Gene Sequence of Nasopharyngeal Exudate Reveals Changes in Key Microbial Communities Associated with Aging.
Candel S, Tyrkalska SD, Pérez-Sanz F, Moreno-Docón A, Esteban Á, Cayuela ML, Mulero V. Candel S, et al. Int J Mol Sci. 2023 Feb 18;24(4):4127. doi: 10.3390/ijms24044127. Int J Mol Sci. 2023. PMID: 36835535 Free PMC article.
References
- Kolter R. A visit to the paediatrician in the not-so-distant future. Environ Microbiol Rep. 2009;1:12–16.
- Lesko LJ, Atkinson AJ., Jr Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: Criteria, validation, strategies. Annu Rev Pharmacol Toxicol. 2001;41:347–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DP2 OD007290/OD/NIH HHS/United States
- DP1 OD000964/OD/NIH HHS/United States
- DE020751/DE/NIDCR NIH HHS/United States
- DP1OD000964/OD/NIH HHS/United States
- P30 DE020751/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources