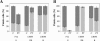Genomic characterization of two Staphylococcus epidermidis bacteriophages with anti-biofilm potential - PubMed (original) (raw)
Genomic characterization of two Staphylococcus epidermidis bacteriophages with anti-biofilm potential
Diana Gutiérrez et al. BMC Genomics. 2012.
Abstract
Background: Staphylococcus epidermidis is a commensal bacterium but can colonize the hospital environment due to its ability to form biofilms favouring adhesion to host tissues, medical devices and increasing resistance to antibiotics. In this context, the use of phages to destroy biofilms is an interesting alternative.
Results: The complete genomes of two Staphylococcus epidermidis bacteriophages, vB_SepiS-phiIPLA5 and vB_SepiS-phiIPLA7, have been analyzed. Their genomes are 43,581 bp and 42,123 bp, and contain 67 and 59 orfs. Bioinformatic analyses enabled the assignment of putative functions to 36 and 29 gene products, respectively, including DNA packaging and morphogenetic proteins, lysis components, and proteins necessary for DNA recombination, regulation, modification and replication. A point mutation in vB_SepiS-phiIPLA5 lysogeny control-associated genes explained its strictly lytic behaviour. Comparative analysis of phi-IPLA5 and phi-IPLA7 genome structure resembled those of S. epidermidis ϕPH15 and ϕCNPH82 phages. A mosaic structure of S. epidermidis prophage genomes was revealed by PCR analysis of three marker genes (integrase, major head protein and holin). Using these genes, high prevalence (73%) of phage DNA in a representative S. epidermidis strain collection consisting of 60 isolates from women with mastitis and healthy women was determined. Putative pectin lyase-like domains detected in virion-associated proteins of both phages could be involved in exopolysaccharide (EPS) depolymerization, as evidenced by both the presence of a clear halo surrounding the phage lysis zone and the phage-mediated biofilm degradation.
Conclusions: Staphylococcus epidermidis bacteriophages, vB_SepiS-phiIPLA5 and vB_SepiS-phiIPLA7, have a mosaic structure similar to other widespread S. epidermidis prophages. Virions of these phages are provided of pectin lyase-like domains, which may be regarded as promising anti-biofilm tools.
Figures
Figure 1
Physical and genetic map of phages phi-IPLA5 (A) and phi-IPLA7 (B). The ORFs are sequentially numbered, indicated by arrows proportional to their lengths and pointing toward their direction of transcription. Some ORFs have been placed below for clarity. The functional modules are indicated on top of the scheme, and the names of several putatively or experimentally identified genes are shown. Putative promoter (P) and terminator (T) sequences are also indicated.
Figure 2
Alignment of the genome of the two S. epidermidis phages phi-IPLA5 and phi-IPLA7 with those of other Staphylococcus phages using the Mauve software. Each block represents a region of the genome sequence that aligned and is homologous to part of another genome. Regions outside blocks lack detectable homology among the input genomes. Inside each block nucleotide sequence similarity is indicated by the height of the colored bars, while regions that are dissimilar are in white. Lines connecting blocks are indicative of homologous regions.
Figure 3
Aminoacid sequence analysis of the virion-associated protein gp18 from phi-IPLA7. A) ClustalW alignment of the predicted binding site. Positions with a single, fully conserved residue are marked with an asterisk; the colon marks the conserved residues between groups of strongly similar properties, and the period marks the residues weakly conserved between groups based on the Gonnet PAM 250 matrix, score <0,5. Highlighted are those residues involved in ligand association. B) Predicted 3D structure of phi-IPLA7 gp18. C) Predicted 3D structure of the domain pectin lyase like (D1) (amino acids 316- 374).
Figure 4
Effect of phages A) phi-IPLA5 and B) phi-IPLA7 on a lawn of S. epidermidis . I) Morphology of phage lytic zone when dropped on different S. epidermidis strains. (S) Sensitive strain; (R) resistant strain; (DS) drop-sensitive strain. II) Bacteria viable number in drop-zone (white), bacteria lawn (black) and phage titre (grey) of phages. Each value correspond with the mean of five different experiments, the standard error is represented by bars.
Figure 5
Killing of S. epidermidis cells forming a biofilm on microtiter wells by phage phi-IPLA7 after incubation for 1 h (A) and 3 h (B). Results are depicted as the percentage of attached cells (dark gray square) and planktonic cells (light gray square) detected in control biofilms treated with SM buffer (C) and in biofilms treated with phage phi-IPLA7 (PT). Each value corresponds with the mean of five different experiments and the standard error is represented by bars
Similar articles
- Isolation and characterization of bacteriophages infecting Staphylococcus epidermidis.
Gutiérrez D, Martínez B, Rodríguez A, García P. Gutiérrez D, et al. Curr Microbiol. 2010 Dec;61(6):601-8. doi: 10.1007/s00284-010-9659-5. Epub 2010 May 7. Curr Microbiol. 2010. PMID: 20449591 - Functional genomic analysis of two Staphylococcus aureus phages isolated from the dairy environment.
García P, Martínez B, Obeso JM, Lavigne R, Lurz R, Rodríguez A. García P, et al. Appl Environ Microbiol. 2009 Dec;75(24):7663-73. doi: 10.1128/AEM.01864-09. Epub 2009 Oct 16. Appl Environ Microbiol. 2009. PMID: 19837832 Free PMC article. - Potential of training of anti-Staphylococcus aureus therapeutic phages against Staphylococcus epidermidis multidrug-resistant isolates is restricted by inter- and intra-sequence type specificity.
Kolenda C, Bonhomme M, Medina M, Pouilly M, Rousseau C, Troesch E, Martins-Simoes P, Stegger M, Verhoeven PO, Laumay F, Laurent F. Kolenda C, et al. mSystems. 2024 Oct 22;9(10):e0085024. doi: 10.1128/msystems.00850-24. Epub 2024 Sep 9. mSystems. 2024. PMID: 39248470 Free PMC article. - Genomics of staphylococcal Twort-like phages--potential therapeutics of the post-antibiotic era.
Łobocka M, Hejnowicz MS, Dąbrowski K, Gozdek A, Kosakowski J, Witkowska M, Ulatowska MI, Weber-Dąbrowska B, Kwiatek M, Parasion S, Gawor J, Kosowska H, Głowacka A. Łobocka M, et al. Adv Virus Res. 2012;83:143-216. doi: 10.1016/B978-0-12-394438-2.00005-0. Adv Virus Res. 2012. PMID: 22748811 Review. - Bacteriophages as Biotechnological Tools.
Elois MA, Silva RD, Pilati GVT, Rodríguez-Lázaro D, Fongaro G. Elois MA, et al. Viruses. 2023 Jan 26;15(2):349. doi: 10.3390/v15020349. Viruses. 2023. PMID: 36851563 Free PMC article. Review.
Cited by
- Isolation and Characterization of New Bacteriophages against Staphylococcal Clinical Isolates from Diabetic Foot Ulcers.
Plumet L, Morsli M, Ahmad-Mansour N, Clavijo-Coppens F, Berry L, Sotto A, Lavigne JP, Costechareyre D, Molle V. Plumet L, et al. Viruses. 2023 Nov 22;15(12):2287. doi: 10.3390/v15122287. Viruses. 2023. PMID: 38140529 Free PMC article. - Translating bacteriophage-derived depolymerases into antibacterial therapeutics: Challenges and prospects.
Wang H, Liu Y, Bai C, Leung SSY. Wang H, et al. Acta Pharm Sin B. 2024 Jan;14(1):155-169. doi: 10.1016/j.apsb.2023.08.017. Epub 2023 Aug 18. Acta Pharm Sin B. 2024. PMID: 38239242 Free PMC article. Review. - The discovery of phiAGATE, a novel phage infecting Bacillus pumilus, leads to new insights into the phylogeny of the subfamily Spounavirinae.
Barylski J, Nowicki G, Goździcka-Józefiak A. Barylski J, et al. PLoS One. 2014 Jan 23;9(1):e86632. doi: 10.1371/journal.pone.0086632. eCollection 2014. PLoS One. 2014. PMID: 24466180 Free PMC article. - Complete Genome of Bacillus megaterium Siphophage Staley.
Hastings WJ, Ritter MA, Chamakura KR, Kuty Everett GF. Hastings WJ, et al. Genome Announc. 2013 Dec 5;1(6):e00864-13. doi: 10.1128/genomeA.00864-13. Genome Announc. 2013. PMID: 24309730 Free PMC article. - Isolation and characterization of bacteriophages from the human skin microbiome that infect Staphylococcus epidermidis.
Valente LG, Pitton M, Fürholz M, Oberhaensli S, Bruggmann R, Leib SL, Jakob SM, Resch G, Que YA, Cameron DR. Valente LG, et al. FEMS Microbes. 2021 Mar 30;2:xtab003. doi: 10.1093/femsmc/xtab003. eCollection 2021. FEMS Microbes. 2021. PMID: 37334235 Free PMC article.
References
- Delgado S, Arroyo R, Jiménez E, Marín ML, Del Campo R, Fernández L, Rodríguez JM. Staphylococcus epidermidis strains isolated from breast milk of women suffering infectious mastitis: potential virulence traits and resistance to antibiotics. BMC Microbiol. 2009;9:82. doi: 10.1186/1471-2180-9-82. - DOI - PMC - PubMed
- Jabbouri S, Sadovskaya I. Characteristics of the biofilm matrix and its role as a possible target for the detection and eradication of Staphylococcus epidermidis associated with medical implant infections. FEMS Immunol Med Microbiol. 2010;59:280–291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources