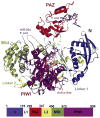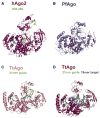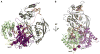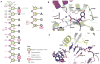The structure of human argonaute-2 in complex with miR-20a - PubMed (original) (raw)
The structure of human argonaute-2 in complex with miR-20a
Elad Elkayam et al. Cell. 2012.
Erratum in
- Cell. 2012 Jul 6;150(1):233
Abstract
Argonaute proteins lie at the heart of the RNA-induced silencing complex (RISC), wherein they use small RNA guides to recognize targets. Initial insight into the architecture of Argonautes came from studies of prokaryotic proteins, revealing a crescent-shaped base made up of the amino-terminal, PAZ, middle, and PIWI domains. The recently reported crystal structure of human Argonaute-2 (hAgo2), the "slicer" in RNA interference, in complex with a mixed population of RNAs derived from insect cells provides insight into the architecture of a eukaryotic Argonaute protein with defined biochemical and biological functions. Here, we report the structure of human Ago2 bound to a physiologically relevant microRNA, microRNA-20a, at 2.2 Å resolution. The miRNA is anchored at both ends by the Mid and PAZ domains and makes several kinks and turns along the binding groove. Interestingly, miRNA binding confers remarkable stability on hAgo2, locking this otherwise flexible enzyme into a stable conformation.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
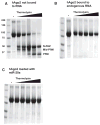
Figure 1. Small RNAs Stabilize hAgo2
(A) Digestion of hAgo2, purified without bound, endogenous small RNAs (Figure 1, lane “not bound to RNA”) with thermolysin resulted in a digestion pattern of typical multidomain proteins. Increasing concentrations of enzyme (indicated by the wedge) released stable protein fragments corresponding to known domains (indicated), as determined by N-terminal sequencing or mass spectrometry. In contrast, treatment of hAgo2 purified along with endogenous SF9 small RNAs was remarkably protease resistant (B). (C) Addition of synthetic miR-20a (1–20) to the fraction shown in (A) conferred stability to protease. See also Figures S1 and S2.
Figure 2. Structure of hAgo2 in Complex with miR-20a
A schematic representation of the hAgo2-miR-20a complex is displayed with each domain and interdomain linker labeled. The N domain is shown in blue, the PAZ domain in red, the Mid domain in green and the PIWI domain in purple. The miRNA is shown in stick representation. The 5′ end of miR-20a is bound to the Mid domain. Nucleotides 2 to 10 track along the RNA binding groove, and nucleotides 17 to 20 are bound to the PAZ domain. The location of the active site is circled. For reference, a bar diagram is given below, color-coded as described, indicating the positions of boundaries between structural domains. Other views of the complex are shown in Figure S2.
Figure 3. Structural Comparison of hAgo2, PfAgo, and Tt Ago
Argonaute proteins across the three kingdoms of life show striking similarities in their overall structures. All structures were superimposed based on their respective PIWI domains and are oriented as in Figure 2. (A) hAgo2-miR-20a complex adopts the most open conformation of all full-length Argonaute structures determined to date. This is compared to the archaeal PfAgo (B), eubacterial TtAgo bound to a 21-mer DNA guide (C), and TtAgo in complex with a 21-mer DNA guide and a 19-mer RNA target (D). See also Figure S4.
Figure 4. Structural Basis of RNA-Induced Stability of hAgo2
(A) hAgo2 is shown in the canonical view as in Figure 2 to highlight interactions of the RNA across all constituent domains of the protein. The two thermolysin cleavage sites at I427 and I577 are shown as red spheres, which are only 10 Å apart and likely require flexibility in the protein for access by the protease. Extensive interactions between the protein and RNA are proposed to “glue” the structure into place, preventing the needed flexibility. (B) An additional view from the MID domain is shown, clarifying the location of the cleavage sites and the path of the RNA.
Figure 5. miR-20a Interactions with hAgo2
(A) A guide to the interactions between miR-20a (the first ten and last four bases that appear unambiguously in the structure) and hAgo2 is shown schematically. Interactions with backbone phosphates, sugars, and bases are indicated according to the key. Interacting residues are color-coded with respect to their domain location as in Figure 3. (B) The site in the Mid domain (light green), which binds the 5′ end of miR-20a is shown. The first miR-20a base (U1) and the terminal monophosphate are displayed along with the interactions they make with hAgo2. Interacting protein side chains are shown in stick representation in atom colors with carbons in white, nitrogens in blue and oxygens in red. The 5′U is shown in atom colors as above, but with carbons in blue and phosphorous in orange. The direction of the RNA chain is shown as a dashed line. The portion of the PIWI domain, which caps the pocket, is shown in purple. (C) The U6-G7 kink and the U9-U10 kink are shown in cartoon form. The L2 Linker is displayed in yellow, the PIWI domain in purple, and the miRNA in stick as in (B). See text for details. See also Figures S5 and S6.
Figure 6. Theoretical Modeling of the Complete miRNA
(A) Nucleotides 11–16 of miR-20a were modeled with guidance from residual electron density (see text). Color coding is as in Figure 3. See also Figure S6. (B) A close-up of the G14-U15 turn is displayed, showing the “proline knuckle” emanating from the N domain on one side and R278 and K280 emanating from the PAZ domain on the other. (C) The sequence of the 20-mer miR-20a is given. Ordered nucleotides are in blue, modeled ones in pale blue. The amino acids in the proline knuckle constriction are shown on both sides of the miRNA in green.
Similar articles
- Crystal Structure of Silkworm PIWI-Clade Argonaute Siwi Bound to piRNA.
Matsumoto N, Nishimasu H, Sakakibara K, Nishida KM, Hirano T, Ishitani R, Siomi H, Siomi MC, Nureki O. Matsumoto N, et al. Cell. 2016 Oct 6;167(2):484-497.e9. doi: 10.1016/j.cell.2016.09.002. Epub 2016 Sep 29. Cell. 2016. PMID: 27693359 - The making of a slicer: activation of human Argonaute-1.
Faehnle CR, Elkayam E, Haase AD, Hannon GJ, Joshua-Tor L. Faehnle CR, et al. Cell Rep. 2013 Jun 27;3(6):1901-9. doi: 10.1016/j.celrep.2013.05.033. Epub 2013 Jun 6. Cell Rep. 2013. PMID: 23746446 Free PMC article. - Understanding the molecular interaction of human argonaute-2 and miR-20a complex: A molecular dynamics approach.
Mallick B, Sharma AR, Lee SS, Chakraborty C. Mallick B, et al. J Cell Biochem. 2019 Dec;120(12):19915-19924. doi: 10.1002/jcb.29300. Epub 2019 Jul 18. J Cell Biochem. 2019. PMID: 31318096 - Eukaryotic Argonautes come into focus.
Kuhn CD, Joshua-Tor L. Kuhn CD, et al. Trends Biochem Sci. 2013 May;38(5):263-71. doi: 10.1016/j.tibs.2013.02.008. Epub 2013 Mar 29. Trends Biochem Sci. 2013. PMID: 23541793 Review. - Why Argonaute is needed to make microRNA target search fast and reliable.
Klein M, Chandradoss SD, Depken M, Joo C. Klein M, et al. Semin Cell Dev Biol. 2017 May;65:20-28. doi: 10.1016/j.semcdb.2016.05.017. Epub 2016 May 26. Semin Cell Dev Biol. 2017. PMID: 27235676 Review.
Cited by
- Stereochemical bias introduced during RNA synthesis modulates the activity of phosphorothioate siRNAs.
Jahns H, Roos M, Imig J, Baumann F, Wang Y, Gilmour R, Hall J. Jahns H, et al. Nat Commun. 2015 Mar 6;6:6317. doi: 10.1038/ncomms7317. Nat Commun. 2015. PMID: 25744034 Free PMC article. - Regulation of Argonaute slicer activity by guide RNA 3' end interactions with the N-terminal lobe.
Hur JK, Zinchenko MK, Djuranovic S, Green R. Hur JK, et al. J Biol Chem. 2013 Mar 15;288(11):7829-7840. doi: 10.1074/jbc.M112.441030. Epub 2013 Jan 17. J Biol Chem. 2013. PMID: 23329841 Free PMC article. - Argonaute proteins: structures and their endonuclease activity.
Jin S, Zhan J, Zhou Y. Jin S, et al. Mol Biol Rep. 2021 May;48(5):4837-4849. doi: 10.1007/s11033-021-06476-w. Epub 2021 Jun 11. Mol Biol Rep. 2021. PMID: 34117606 Review. - Expression and Functional Analysis of the Argonaute Protein of Thermus thermophilus (TtAgo) in E. coli BL21(DE3).
Xing J, Ma L, Cheng X, Ma J, Wang R, Xu K, Mymryk JS, Zhang Z. Xing J, et al. Biomolecules. 2021 Mar 31;11(4):524. doi: 10.3390/biom11040524. Biomolecules. 2021. PMID: 33807395 Free PMC article. - A spontaneous thermo-sensitive female sterility mutation in rice enables fully mechanized hybrid breeding.
Li H, You C, Yoshikawa M, Yang X, Gu H, Li C, Cui J, Chen X, Ye N, Zhang J, Wang G. Li H, et al. Cell Res. 2022 Oct;32(10):931-945. doi: 10.1038/s41422-022-00711-0. Epub 2022 Sep 6. Cell Res. 2022. PMID: 36068348 Free PMC article.
References
- Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr. 1996;52:30–42. - PubMed
- Bieniossek C, Richmond TJ, Berger I. MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr Protoc Protein Sci. 2008;Chapter 5(Unit 5.20) - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM062534/GM/NIGMS NIH HHS/United States
- R01 GM062534/GM/NIGMS NIH HHS/United States
- P30 EB009998/EB/NIBIB NIH HHS/United States
- P30 CA045508/CA/NCI NIH HHS/United States
- R37 GM062534/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials