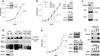BAP1 loss defines a new class of renal cell carcinoma - PubMed (original) (raw)
. 2012 Jun 10;44(7):751-9.
doi: 10.1038/ng.2323.
Silvia Vega-Rubín-de-Celis, Arnold Liao, Nan Leng, Andrea Pavía-Jiménez, Shanshan Wang, Toshinari Yamasaki, Leah Zhrebker, Sharanya Sivanand, Patrick Spence, Lisa Kinch, Tina Hambuch, Suneer Jain, Yair Lotan, Vitaly Margulis, Arthur I Sagalowsky, Pia Banerji Summerour, Wareef Kabbani, S W Wendy Wong, Nick Grishin, Marc Laurent, Xian-Jin Xie, Christian D Haudenschild, Mark T Ross, David R Bentley, Payal Kapur, James Brugarolas
Affiliations
- PMID: 22683710
- PMCID: PMC3788680
- DOI: 10.1038/ng.2323
BAP1 loss defines a new class of renal cell carcinoma
Samuel Peña-Llopis et al. Nat Genet. 2012.
Erratum in
- Nat Genet. 2012 Sep;44(9):1072
Abstract
The molecular pathogenesis of renal cell carcinoma (RCC) is poorly understood. Whole-genome and exome sequencing followed by innovative tumorgraft analyses (to accurately determine mutant allele ratios) identified several putative two-hit tumor suppressor genes, including BAP1. The BAP1 protein, a nuclear deubiquitinase, is inactivated in 15% of clear cell RCCs. BAP1 cofractionates with and binds to HCF-1 in tumorgrafts. Mutations disrupting the HCF-1 binding motif impair BAP1-mediated suppression of cell proliferation but not deubiquitination of monoubiquitinated histone 2A lysine 119 (H2AK119ub1). BAP1 loss sensitizes RCC cells in vitro to genotoxic stress. Notably, mutations in BAP1 and PBRM1 anticorrelate in tumors (P = 3 × 10(-5)), [corrected] and combined loss of BAP1 and PBRM1 in a few RCCs was associated with rhabdoid features (q = 0.0007). BAP1 and PBRM1 regulate seemingly different gene expression programs, and BAP1 loss was associated with high tumor grade (q = 0.0005). Our results establish the foundation for an integrated pathological and molecular genetic classification of RCC, paving the way for subtype-specific treatments exploiting genetic vulnerabilities.
Figures
Fig. 1. Integrative mutation and DNA copy-number analyses in a tumor and tumorgraft from the index subject
(a) Representative capillary sequencing chromatograms of normal (N), patient’s tumor (T), and tumorgraft (TG) illustrating different examples of mutant allele enrichment in the tumorgraft. Arrowheads indicate mutations. (b) Allele specific (ASCN) and paired (PCN) copy number representation of high-density SNP array data incorporating the estimated position of mutated genes. Green, paired copy numbers. Red and blue, maximum and minimum copy numbers for each heterozygous SNP.
Fig. 2. BAP1 is a tumor suppressor in ccRCC
(a) Schematic of BAP1 with mutations (UCH, ubiquitin C-terminal hydrolase domain; HBM, HCF-1 binding motif; BRCA1, [putative] BRCA1 interacting domain; ULD, Uch37-like domain; NLS, nuclear localization signal; I, insertion; Δ, deletion; †, missense; *, non-sense; S, splice site; *L, stop codon loss. (b) Structural model of BAP1 UCH domain (purple) and ULD tail (green) superimposed on a template DUB (not shown) bound to ubiquitin (cyan); structural elements that alter upon ubiquitin binding are colored salmon. Left, cartoon of BAP1 model. Right, surface representation of BAP1 highlighting the positions of RCC alterations on interaction surfaces. Left inset, enlarged view of the DUB active site. Right inset, enlarged view of the Gly13, interaction with an aromatic residue in the ULD tail (dots indicate interaction radius). (c) Western blot of extracts from tumors with defined BAP1 mutation and chromosome 3p status. 769-P cells transfected with either an empty vector (EV) or wild-type (WT) BAP1 were used as controls. PPIB, cyclophilin B is shown as a loading control. Arrow indicates BAP1. (d) Representative IHC of tumors and tumorgrafts positive or negative for nuclear BAP1. Scale bar, 50 μm. Open arrow, tumor cells; simple arrow, endothelial cells and lymphocytes, which express BAP1 and serve as internal controls.
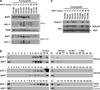
Fig. 3. HCF-1-dependent suppression of cell proliferation by BAP1
(a) Proliferation curves of empty vector (EV) and BAP1 reconstituted 769-P cells with inset showing BAP1 western blot. (b) Proliferation curves of 769-P cells stably expressing an shRNA targeting endogenous (mutant) BAP1 (shBAP1) or a vector control (shCtrl) and in addition wild-type BAP1 (BAP1), BAP1Y33D (Y33D) or empty vector (EV). Western blot of cells transduced with shRNA targeting endogenous mutant BAP1 (A and B) or vector control (shCtrl) and transduced with expression vectors as indicated. (c) Western blot of partially purified histone fractions of 769-P cells reconstituted with an empty vector (EV) or wild-type BAP1 (BAP1). (d) Western blot of input (cytosolic [Cy] or nuclear [Nu] fractions) as well as immunoprecipitates (from nuclear fractions) and corresponding flow-through from empty vector (−) or wild-type BAP1 (+) expressing cells. Short and long exposures are indicated. Both ectopically expressed epitope tagged wild-type as well as endogenous mutant BAP1 bind HCF-1. (e) Proliferation curves of 769-P cells depleted of endogenous BAP1 shRNA and reconstituted with an empty vector (EV), wild-type BAP1 (WT) or HCF-1 binding motif mutant (HBM). Western blot from input as well as BAP1 immunoprecipitates. (f) Western blot of partially purified histone fractions or cell lysates from 769-P cells depleted of endogenous_BAP1_ and transduced with an empty vector (EV), wild-type BAP1 (WT), an HCF-1 binding motif mutant (HBM), or BAP1Y33D (Y33D). Error bars represent SEM (_n_=3). *,p<0.05; **, p<0.01.
Fig. 4. BAP1 binds to and elutes with HCF-1 in tumorgrafts
(a) Western blot from inputs as well as BAP1 immunoprecipitates (BAP1-IP) from the indicated tumorgrafts. wt, wild type; M, mutant. (b) Western blot of TCA precipitated gel-filtration fractions of tumorgrafts either wild type for BAP1 (TG143 and TG144) or mutant (TG26). Ct, Control lysate from 769-P cells. (c) Western blot of partially purified histone fractions from tumorgrafts with the indicated BAP1 status.
Fig. 5. Loss of BAP1 and PBRM1 sets foundation for molecular genetic classification of ccRCC
(a) Representative H&E and immunohistochemistry (IHC) images of tumors with loss of BAP1, PBRM1, or both. Scale bar, 50 µm; 10 µm for inset. Open arrows, tumor cells; simple arrows, stroma/inflammatory cells; filled arrow, rhabdoid tumor cell. (b) Pie chart of the distribution of ccRCC subtypes. (c) Heatmap of statistically significant probes distinguishing _BAP1_- and_PBRM1_-deficient tumors/tumorgrafts vs. wild type. Expression of the same probes in renal cortex included as a reference. The full data set is provided in Supplementary Data 4. (d) Venn diagram illustrating the overlap in BAP1 and PBRM1 gene expression signatures with associated global pathway analyses.
Similar articles
- Loss of PBRM1 and BAP1 expression is less common in non-clear cell renal cell carcinoma than in clear cell renal cell carcinoma.
Ho TH, Kapur P, Joseph RW, Serie DJ, Eckel-Passow JE, Parasramka M, Cheville JC, Wu KJ, Frenkel E, Rakheja D, Stefanius K, Brugarolas J, Parker AS. Ho TH, et al. Urol Oncol. 2015 Jan;33(1):23.e9-23.e14. doi: 10.1016/j.urolonc.2014.10.014. Epub 2014 Nov 24. Urol Oncol. 2015. PMID: 25465300 Free PMC article. - Genomic profiling of the genes on chromosome 3p in sporadic clear cell renal cell carcinoma.
Togo Y, Yoshikawa Y, Suzuki T, Nakano Y, Kanematsu A, Zozumi M, Nojima M, Hirota S, Yamamoto S, Hashimoto-Tamaoki T. Togo Y, et al. Int J Oncol. 2016 Apr;48(4):1571-80. doi: 10.3892/ijo.2016.3395. Epub 2016 Feb 17. Int J Oncol. 2016. PMID: 26891804 - Exploration of Morphological Features of Clear Cell Renal Cell Carcinoma With PBRM1, SETD2, BAP1, or KDM5C Mutations.
Liu Y, Li Y, Xu H, Zhou L, Yang X, Wang C. Liu Y, et al. Int J Surg Pathol. 2023 Dec;31(8):1485-1494. doi: 10.1177/10668969231157317. Epub 2023 Mar 13. Int J Surg Pathol. 2023. PMID: 36911986 - BAP1, PBRM1 and SETD2 in clear-cell renal cell carcinoma: molecular diagnostics and possible targets for personalized therapies.
Piva F, Santoni M, Matrana MR, Satti S, Giulietti M, Occhipinti G, Massari F, Cheng L, Lopez-Beltran A, Scarpelli M, Principato G, Cascinu S, Montironi R. Piva F, et al. Expert Rev Mol Diagn. 2015;15(9):1201-10. doi: 10.1586/14737159.2015.1068122. Epub 2015 Jul 11. Expert Rev Mol Diagn. 2015. PMID: 26166446 Review. - The expanding role of BAP1 in clear cell renal cell carcinoma.
Kapur P, Rajaram S, Brugarolas J. Kapur P, et al. Hum Pathol. 2023 Mar;133:22-31. doi: 10.1016/j.humpath.2022.07.022. Epub 2022 Aug 4. Hum Pathol. 2023. PMID: 35932824 Free PMC article. Review.
Cited by
- ATF4/NUPR1 axis promotes cancer cell survival and mediates immunosuppression in clear cell renal cell carcinoma.
Lu Y, Chen W, Xuan Y, Li X, Wu S, Wang H, Guo T, Wang C, Tian S, Li H, Lai D, Zhao W, Huang X, Zhao X, Wang B, Zhang X, Li H, Huang Y, Ma X. Lu Y, et al. Discov Oncol. 2024 Oct 31;15(1):607. doi: 10.1007/s12672-024-01485-0. Discov Oncol. 2024. PMID: 39480570 Free PMC article. - Distinct molecular profiles and shared drug vulnerabilities in pancreatic metastases of renal cell carcinoma.
Roos-Mattila M, Kallio P, Luck TJ, Polso M, Kumari R, Mikkonen P, Välimäki K, Malmstedt M, Ellonen P, Pellinen T, Heckman CA, Mustonen H, Puolakkainen PA, Alitalo K, Kallioniemi O, Mirtti T, Rannikko AS, Pietiäinen VM, Seppänen HE. Roos-Mattila M, et al. Commun Biol. 2024 Oct 20;7(1):1355. doi: 10.1038/s42003-024-07004-9. Commun Biol. 2024. PMID: 39427059 Free PMC article. - Screening of differential gene expression patterns through survival analysis for diagnosis, prognosis and therapies of clear cell renal cell carcinoma.
Ajadee A, Mahmud S, Hossain MB, Ahmmed R, Ali MA, Reza MS, Sarker SK, Mollah MNH. Ajadee A, et al. PLoS One. 2024 Sep 30;19(9):e0310843. doi: 10.1371/journal.pone.0310843. eCollection 2024. PLoS One. 2024. PMID: 39348357 Free PMC article. - In Silico Exploration of AHR-HIF Pathway Interplay: Implications for Therapeutic Targeting in ccRCC.
Gregoris F, Minervini G, Tosatto SCE. Gregoris F, et al. Genes (Basel). 2024 Sep 5;15(9):1167. doi: 10.3390/genes15091167. Genes (Basel). 2024. PMID: 39336758 Free PMC article. - Tumor suppressor BAP1 suppresses disulfidptosis through the regulation of SLC7A11 and NADPH levels.
Wang J, Wang M, Wu S, Zhu Y, Fan K, Chen Y, Xiao Z, Chen J, Tu K, Huang D, Zhang Y, Xu Q. Wang J, et al. Oncogenesis. 2024 Sep 12;13(1):31. doi: 10.1038/s41389-024-00535-0. Oncogenesis. 2024. PMID: 39266549 Free PMC article.
References
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. - PubMed
- Baldewijns MM, et al. Genetics and epigenetics of renal cell cancer. Biochim Biophys Acta. 2008;1785:133–155. - PubMed
- Brugarolas J. Renal-cell carcinoma--molecular pathways and therapies. N Engl J Med. 2007;356:185–187. - PubMed
- Latif F, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. - PubMed
- Gnarra JR, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous