The intermediate filament protein, vimentin, is a regulator of NOD2 activity - PubMed (original) (raw)
The intermediate filament protein, vimentin, is a regulator of NOD2 activity
Craig Stevens et al. Gut. 2013 May.
Erratum in
- Gut. 2013 May;62(5):707
Abstract
Objective: Mutations in the nucleotide-binding oligomerisation domain-containing protein 2 (NOD2) gene remain the strongest genetic determinants for Crohn's disease (CD). Having previously identified vimentin as a novel NOD2-interacting protein, the authors aimed to investigate the regulatory effects of vimentin on NOD2 function and the association of variants in Vim with CD susceptibility.
Design: Coimmunoprecipitation, fluorescent microscopy and fractionation were used to confirm the interaction between NOD2 and vimentin. HEK293 cells stably expressing wild-type NOD2 or a NOD2 frameshift variant (L1007fs) and SW480 colonic epithelial cells were used alongside the vimentin inhibitor, withaferin A (WFA), to assess effects on NOD2 function using the nuclear factor-kappaB (NF-κB) reporter gene, green fluorescent protein-LC3-based autophagy, and bacterial gentamicin protection assays. International genome-wide association meta-analysis data were used to test for associations of single-nucleotide polymorphisms in Vim with CD susceptibility.
Results: The leucine-rich repeat domain of NOD2 contained the elements required for vimentin binding; CD-associated polymorphisms disrupted this interaction. NOD2 and vimentin colocalised at the cell plasma membrane, and cytosolic mislocalisation of the L1007fs and R702W variants correlated with an inability to interact with vimentin. Use of WFA demonstrated that vimentin was required for NOD2-dependent NF-κB activation and muramyl dipeptide-induced autophagy induction, and that NOD2 and vimentin regulated the invasion and survival properties of a CD-associated adherent-invasive Escherichia coli strain. Genetic analysis revealed an association signal across the haplotype block containing Vim.
Conclusion: Vimentin is an important regulator of NOD2 function and a potential novel therapeutic target in the treatment of CD. In addition, Vim is a candidate susceptibility gene for CD, supporting the functional data.
Figures
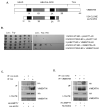
Figure 1. Identification of vimentin as a NOD2 interacting protein
(a) Schematic showing the structure of vimentin and the vimentin clone identified as binding to NOD2 in our yeast-two-hybrid assay. (b) Yeast strain AH109 was co-transformed with a plasmid containing the yeast expression vector pGADT7-AD, full-length vimentin cDNA, wild-type NOD2 or CD-associated variant NOD2-R702W as shown. Co-transformants were selected on (Leu-Trp-) plates and spotted onto selective media (His-Leu-Trp-). (Dashed line indicates where irrelevant lanes have been removed). (c) SW480 cells were transfected with HA-NOD2 or HA-empty vector and extracts immunoprecipitated with HA-11 specific antibodies or non-specific antibody as control. Precipitates were immunoblotted for associated vimentin and NOD2, and direct lysates with HA-11 and vimentin antibodies. (d) SW480 cells were transfected with HA-NOD2 or HA-empty vector and cell extracts immunoprecipitated with vimentin specific antibodies or non-specific antibody as control. Precipitates were immunoblotted for associated NOD2 and vimentin, and direct lysates with HA-11 and vimentin antibodies.
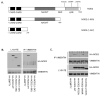
Figure 2. The LRR domain contains the major determinant for binding to vimentin
(a) Schematic showing the structure of NOD2 and the location of CD-associated polymorphisms. The NOD2 deletion mutants lacking the LRR domain or lacking both the LRR and NACHT (NAIP, CIITA, HET-E and TP1) domains generated by site-directed mutagenesis are also shown. (b) HA-NOD2 or the deletion mutants were transfected into HEK293 cells and cell lysates immunoprecipitated with vimentin specific antibodies. Precipitates were immunoblotted for associated NOD2 proteins and vimentin, and direct lysates with HA-11 and vimentin antibodies. (c) HEK293 cells were transfected with HA-NOD2, HA-NOD2-R702W, HA-NOD2-G908R or HA-NOD2-L1007fs variants, and cell lysates immunoprecipitated with vimentin specific antibodies. Precipitates were immunoblotted for associated NOD2 and vimentin, and direct lysates with HA-11 and vimentin antibodies.
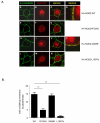
Figure 3. Subcellular localisation of CD-associated NOD2 variants
(a) HEK293 cells were transfected with HA-NOD2, HA-NOD2-R702W, HA-NOD2 - G908R or HA-NOD2-L1007fs variants and immunostained with E-cadherin specific antibodies (green) and exogenous NOD2 with HA-11 antibodies (red). Merged images demonstrate areas where NOD2 and E-cadherin co-localise (yellow) and are highlighted in HA-NOD2 and HA-NOD2-G908R cells (panels d,k). (b) Quantification of NOD2 membrane localisation in Figure 2A. Twenty cells from three separate fields of view were evaluated for membrane localized NOD2. *** = P<0.001. (c) HEK293 cells were transfected with HA-NOD2, HA-NOD2-R702W, HA-NOD2 - G908R or HA-NOD2-L1007fs variants. Following transfection, proteins were separated into specific subcellular fractionations. Each fraction was immunoblotted for NOD2 proteins, vimentin, GAPDH, E-cadherin and Histone H1. (d) Relative protein membrane fraction of HA-NOD2 (i), HA-NOD2-R702W (ii), HA-NOD2-G908R (iii) and HA-NOD2-L1007fs (iv) in relation to the cytosolic fraction using densitometry. Cyt, cytosol; Mem, membrane. (e) HEK293 cells were transfected with HA-NOD2 and immunostained with HA-11 antibodies (red) vimentin-specific antibodies (green). Areas where NOD2 and vimentin co-localise are highlighted and appear as yellow.
Figure 3. Subcellular localisation of CD-associated NOD2 variants
(a) HEK293 cells were transfected with HA-NOD2, HA-NOD2-R702W, HA-NOD2 - G908R or HA-NOD2-L1007fs variants and immunostained with E-cadherin specific antibodies (green) and exogenous NOD2 with HA-11 antibodies (red). Merged images demonstrate areas where NOD2 and E-cadherin co-localise (yellow) and are highlighted in HA-NOD2 and HA-NOD2-G908R cells (panels d,k). (b) Quantification of NOD2 membrane localisation in Figure 2A. Twenty cells from three separate fields of view were evaluated for membrane localized NOD2. *** = P<0.001. (c) HEK293 cells were transfected with HA-NOD2, HA-NOD2-R702W, HA-NOD2 - G908R or HA-NOD2-L1007fs variants. Following transfection, proteins were separated into specific subcellular fractionations. Each fraction was immunoblotted for NOD2 proteins, vimentin, GAPDH, E-cadherin and Histone H1. (d) Relative protein membrane fraction of HA-NOD2 (i), HA-NOD2-R702W (ii), HA-NOD2-G908R (iii) and HA-NOD2-L1007fs (iv) in relation to the cytosolic fraction using densitometry. Cyt, cytosol; Mem, membrane. (e) HEK293 cells were transfected with HA-NOD2 and immunostained with HA-11 antibodies (red) vimentin-specific antibodies (green). Areas where NOD2 and vimentin co-localise are highlighted and appear as yellow.
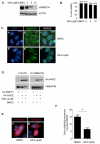
Figure 4. WFA inhibits vimentin and relocalises NOD2 to the cytosol
(a) HEK293 cells were treated with WFA or DMSO control as indicated for 2h. Cell extracts were then immunoblotted for vimentin and actin. (b) The viability of cells treated with WFA or DMSO control for 2h at the indicated concentrations was evaluated using trypan blue exclusion. (c) HEK293 cells were treated with WFA (2μM) or DMSO control for 2h. Cells were then immunostained with vimentin-specific antibodies and co-stained with DAPI to detect nuclei. (d) HEK293 cells transfected with HA-NOD2 were treated with WFA (2μM) or DMSO control for 2h, and cell lysates immunoprecipitated with vimentin specific antibodies. Precipitates were immunoblotted for associated NOD2 and vimentin, and direct lysates with HA-11 and vimentin antibodies. (e) HEK293 cells were transfected with HA-NOD2. Cells were then treated with WFA (2μM) or DMSO control for 2h. Following treatment cells were immunostained with HA-11 antibodies and co-stained with DAPI to detect nuclei. (f) Quantification of NOD2 membrane localisation in Figure 4E. Twenty cells from three separate fields of view were evaluated for membrane localised NOD2. *** = P<0.001.
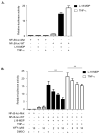
Figure 5. WFA inhibits NOD2-dependent NF-κB signaling
(a) SW480 cells were transfected with NF-κB-luciferase wild-type (NF-κB-luc-WT) or mutant with NF-κB binding sites deleted (NF-κB-luc-Mut) (200ng) and the internal control renilla (100ng). After 16-18h cells were treated for 6h with L18-MDP (1μg/ml) or for 2h with TNF-α (50ng/ml) and harvested for luciferase assays. The data are derived from triplicate readings. (b) SW480 cells were transfected with NF-κB-luciferase wild-type (NF-κB-luc-WT) or mutant with NF-κB binding sites deleted (NF-κB-luc-Mut) (200ng) and the internal control renilla (100ng). After 16-18h cells were treated for 6h with L18-MDP (1μg/ml) or for 2h with TNF-α (50ng/ml) in the presence of WFA (2h) or DMSO control (2h) and harvested for luciferase assays. The data are derived from triplicate readings. * = P<0.05, ns = not significant.
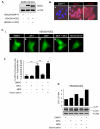
Figure 6. WFA inhibits NOD2-dependent autophagy
(a) HEK293 cells stably expressing FLAG-EMPTY, FLAG-NOD2 or FLAG-NOD2-L1007fs were generated. Cell extracts were immunoblotted with NOD2-specific antibodies. (b) HEK293 cells stably expressing FLAG-EMPTY, FLAG-NOD2 or FLAG-NOD2-L1007fs were immunostained with NOD2-specific antibodies. (c) and (e) HEK293 cells stably expressing FLAG-NOD2 (c) or FLAG-NOD2-L1007fs (e) were transfected to express GFP-LC3. Cells were pre-treated for 30min with WFA (2μM) before treatment for 8h with L18-MDP (50μg/ml). Following treatment the percentage of cells exhibiting greater than five distinct LC3 punctate autophagosomes per cell was determined by fluorescence microscopy (i) and quantified in (ii). GFP-LC3 positive cells in 5 separate fields of view were counted for each sample. ** = P<0.01 (d) and (f) HEK293 cells stably expressing FLAG-NOD2 (d) or FLAG-L1007fs (f) were pre-treated for 30min with WFA (2μM) before treatment for 8h with L18-MDP (50μg/ml). Cell extracts were immunoblotted for LC3 and actin. The ratio of LC3-II: LC3-I was measured by densitometry. S/S, serum starved.
Figure 6. WFA inhibits NOD2-dependent autophagy
(a) HEK293 cells stably expressing FLAG-EMPTY, FLAG-NOD2 or FLAG-NOD2-L1007fs were generated. Cell extracts were immunoblotted with NOD2-specific antibodies. (b) HEK293 cells stably expressing FLAG-EMPTY, FLAG-NOD2 or FLAG-NOD2-L1007fs were immunostained with NOD2-specific antibodies. (c) and (e) HEK293 cells stably expressing FLAG-NOD2 (c) or FLAG-NOD2-L1007fs (e) were transfected to express GFP-LC3. Cells were pre-treated for 30min with WFA (2μM) before treatment for 8h with L18-MDP (50μg/ml). Following treatment the percentage of cells exhibiting greater than five distinct LC3 punctate autophagosomes per cell was determined by fluorescence microscopy (i) and quantified in (ii). GFP-LC3 positive cells in 5 separate fields of view were counted for each sample. ** = P<0.01 (d) and (f) HEK293 cells stably expressing FLAG-NOD2 (d) or FLAG-L1007fs (f) were pre-treated for 30min with WFA (2μM) before treatment for 8h with L18-MDP (50μg/ml). Cell extracts were immunoblotted for LC3 and actin. The ratio of LC3-II: LC3-I was measured by densitometry. S/S, serum starved.
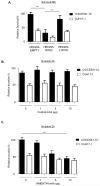
Figure 7. NOD2 and vimentin regulate the invasion and survival of a CD-associated AIEC
(a) Bacterial survival was evaluated in HEK293 cells stably expressing FLAG-EMPTY, FLAG-NOD2 or FLAG-NOD2-L1007fs by gentamicin protection assay after 24h using the E.coli strain CUICD541-10 or CUH17-1. ** = P<0.01, * = P<0.05. (b) Suppression of E.coli strain CUICD541-10 or CUH17-1 invasion of HEK293 cells by pre-incubation of host cells with non-specific control antibodies, (c) antibodies that specifically block vimentin, (d) WFA or DMSO control, was assessed by gentamicin protection assay after 2h. *** = P<0.001. (e) To assess the effect of WFA on E.coli strain CUICD541-10 survival, bacteria were allowed to invade HEK293 cells stably expressing FLAG-EMPTY or FLAG-NOD2 prior to treatment with WFA or DMSO control. The effect of NOD2 and WFA on the survival of CUICD541-10 was assessed by gentamicin protection assay after 24h. ** = P<0.01, * = P<0.05.
Figure 7. NOD2 and vimentin regulate the invasion and survival of a CD-associated AIEC
(a) Bacterial survival was evaluated in HEK293 cells stably expressing FLAG-EMPTY, FLAG-NOD2 or FLAG-NOD2-L1007fs by gentamicin protection assay after 24h using the E.coli strain CUICD541-10 or CUH17-1. ** = P<0.01, * = P<0.05. (b) Suppression of E.coli strain CUICD541-10 or CUH17-1 invasion of HEK293 cells by pre-incubation of host cells with non-specific control antibodies, (c) antibodies that specifically block vimentin, (d) WFA or DMSO control, was assessed by gentamicin protection assay after 2h. *** = P<0.001. (e) To assess the effect of WFA on E.coli strain CUICD541-10 survival, bacteria were allowed to invade HEK293 cells stably expressing FLAG-EMPTY or FLAG-NOD2 prior to treatment with WFA or DMSO control. The effect of NOD2 and WFA on the survival of CUICD541-10 was assessed by gentamicin protection assay after 24h. ** = P<0.01, * = P<0.05.
Figure 8. Results of a meta-analysis of 7 CD genome-wide association studies imputed with the 1000 genomes reference set
Scatterplot showing −logP values in the 163kb region of Vim for 965 single nucleotide polymorphisms. Vertical dotted lines represent the boundaries of the Vim gene and solid lines the limits of each haplotype block. The −logP value corresponding to P<0.05 is represented by the horizontal (red) dotted line.
Similar articles
- TLE1 modifies the effects of NOD2 in the pathogenesis of Crohn's disease.
Nimmo ER, Stevens C, Phillips AM, Smith A, Drummond HE, Noble CL, Quail M, Davies G, Aldhous MC, Wilson DC, Satsangi J. Nimmo ER, et al. Gastroenterology. 2011 Sep;141(3):972-981.e1-2. doi: 10.1053/j.gastro.2011.05.043. Epub 2011 May 27. Gastroenterology. 2011. PMID: 21699783 - ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis.
Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. Homer CR, et al. Gastroenterology. 2010 Nov;139(5):1630-41, 1641.e1-2. doi: 10.1053/j.gastro.2010.07.006. Epub 2010 Jul 14. Gastroenterology. 2010. PMID: 20637199 Free PMC article. - A role for vimentin in Crohn disease.
Henderson P, Wilson DC, Satsangi J, Stevens C. Henderson P, et al. Autophagy. 2012 Nov;8(11):1695-6. doi: 10.4161/auto.21690. Epub 2012 Aug 28. Autophagy. 2012. PMID: 22929019 Free PMC article. - Nucleotide-binding oligomerization domain containing 2: structure, function, and diseases.
Yao Q. Yao Q. Semin Arthritis Rheum. 2013 Aug;43(1):125-30. doi: 10.1016/j.semarthrit.2012.12.005. Epub 2013 Jan 24. Semin Arthritis Rheum. 2013. PMID: 23352252 Review. - The complex interplay of NOD-like receptors and the autophagy machinery in the pathophysiology of Crohn disease.
Billmann-Born S, Lipinski S, Böck J, Till A, Rosenstiel P, Schreiber S. Billmann-Born S, et al. Eur J Cell Biol. 2011 Jun-Jul;90(6-7):593-602. doi: 10.1016/j.ejcb.2010.10.015. Epub 2010 Dec 10. Eur J Cell Biol. 2011. PMID: 21146253 Review.
Cited by
- NOD1, NOD2, and NLRC5 Receptors in Antiviral and Antimycobacterial Immunity.
Godkowicz M, Druszczyńska M. Godkowicz M, et al. Vaccines (Basel). 2022 Sep 7;10(9):1487. doi: 10.3390/vaccines10091487. Vaccines (Basel). 2022. PMID: 36146565 Free PMC article. Review. - Human Vimentin Layers on Solid Substrates: Adsorption Kinetics and Corona Formation Investigations.
Wasilewska M, Żeliszewska P, Pogoda K, Deptuła P, Bucki R, Adamczyk Z. Wasilewska M, et al. Biomacromolecules. 2022 Aug 8;23(8):3308-3317. doi: 10.1021/acs.biomac.2c00415. Epub 2022 Jul 13. Biomacromolecules. 2022. PMID: 35829774 Free PMC article. - The role of intermediate filaments in maintaining integrity and function of intestinal epithelial cells after massive bowel resection in a rat.
Sukhotnik I, Shahar YB, Pollak Y, Dorfman T, Shefer HK, Assi ZE, Mor-Vaknin N, Coran AG. Sukhotnik I, et al. Pediatr Surg Int. 2018 Feb;34(2):217-225. doi: 10.1007/s00383-017-4192-2. Epub 2017 Oct 17. Pediatr Surg Int. 2018. PMID: 29043445 - Vimentin Regulates Chemokine Expression and NOD2 Activation in Brain Endothelium during Group B Streptococcal Infection.
Villarreal R, Manzer HS, Keestra-Gounder AM, Doran KS. Villarreal R, et al. Infect Immun. 2021 Nov 16;89(12):e0034021. doi: 10.1128/IAI.00340-21. Epub 2021 Sep 7. Infect Immun. 2021. PMID: 34491787 Free PMC article. - The molecular biophysics of extracellular vimentin and its role in pathogen-host interactions.
Parvanian S, Coelho-Rato LS, Eriksson JE, Patteson AE. Parvanian S, et al. Curr Opin Cell Biol. 2023 Dec;85:102233. doi: 10.1016/j.ceb.2023.102233. Epub 2023 Sep 5. Curr Opin Cell Biol. 2023. PMID: 37677998 Free PMC article. Review.
References
- Lees CW, Barrett JC, Parkes M, et al. New IBD genetics: common pathways with other diseases. Gut. 2011;60(12):1739–53. - PubMed
- Henderson P, Van Limbergen J, Wilson DC, et al. Genetics of childhood-onset inflammatory bowel disease. Inflamm Bowel Dis. 2011;17(1):346–61. - PubMed
- Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127(2):412–21. - PubMed
- Girardin SE, Boneca IG, Viala J, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278(11):8869–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CZB/4/540/CSO_/Chief Scientist Office/United Kingdom
- ETM/137/CSO_/Chief Scientist Office/United Kingdom
- CZB/4/585/CSO_/Chief Scientist Office/United Kingdom
- G1002033/MRC_/Medical Research Council/United Kingdom
- G0800675/MRC_/Medical Research Council/United Kingdom
- G0600329/MRC_/Medical Research Council/United Kingdom
- G0800759/MRC_/Medical Research Council/United Kingdom
- ETM/75/CSO_/Chief Scientist Office/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous