Synthesis, release, and recapture of compatible solute proline by osmotically stressed Bacillus subtilis cells - PubMed (original) (raw)
Synthesis, release, and recapture of compatible solute proline by osmotically stressed Bacillus subtilis cells
Tamara Hoffmann et al. Appl Environ Microbiol. 2012 Aug.
Abstract
Bacillus subtilis synthesizes large amounts of the compatible solute proline as a cellular defense against high osmolarity to ensure a physiologically appropriate level of hydration of the cytoplasm and turgor. It also imports proline for this purpose via the osmotically inducible OpuE transport system. Unexpectedly, an opuE mutant was at a strong growth disadvantage in high-salinity minimal media lacking proline. Appreciable amounts of proline were detected in the culture supernatant of the opuE mutant strain, and they rose concomitantly with increases in the external salinity. We found that the intracellular proline pool of severely salinity-stressed cells of the opuE mutant was considerably lower than that of its opuE(+) parent strain. This loss of proline into the medium and the resulting decrease in the intracellular proline content provide a rational explanation for the observed salt-sensitive growth phenotype of cells lacking OpuE. None of the known MscL- and MscS-type mechanosensitive channels of B. subtilis participated in the release of proline under permanently imposed high-salinity growth conditions. The data reported here show that the OpuE transporter not only possesses the previously reported role for the scavenging of exogenously provided proline as an osmoprotectant but also functions as a physiologically highly important recapturing device for proline that is synthesized de novo and subsequently released by salt-stressed B. subtilis cells. The wider implications of our findings for the retention of compatible solutes by osmotically challenged microorganisms and the roles of uptake systems for compatible solutes are considered.
Figures
Fig 1
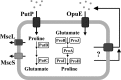
Synthesis, uptake, release, and consumption of proline in B. subtilis. Synthesis of proline for anabolic purposes is carried out by the ProB, ProA, and ProI enzymes; their structural genes (proBA, proI) are induced by proline starvation via a proline-sensing T-box regulatory mechanism (11). The ProJ, ProA, and ProH enzymes carry out synthesis of proline as an osmoprotectant; expression of the proHJ operon is induced by increases in the external salinity but that of the proA gene is not induced in response to this stimulus (10). Uptake of proline as an osmoprotectant is mediated by the OpuE transporter (56), whose structural gene is induced in cells grown at high salinity (47). Use of proline as a carbon or nitrogen source is catalyzed via import through the OpuE-related PutP transporter and subsequent degradation to glutamate via the PutB and PutC enzymes; expression of the putBCP operon is induced by the presence of proline in the growth medium (39). The excretion of proline is depicted either as a carrier/transporter-mediated process or through passive passage across the cytoplasmic membrane. MscL and MscS are mechanosensitive channels whose transient gating protects osmotically down-shocked cells from lysis (24, 58).
Fig 2
Effects of high salinity on the growth of the opuE mutant strain BLOB9 and its parent strain JH642. Cells of strain JH642 (opuE+) (●) and BLOB9 [Δ(opuE::tet)_1_] (▲) were grown at 37°C in SMM (A) or SMM with 1.2 M NaCl (B). Growth of strain JSB8 [Δ(proHJ::tet)_1_] (■) was also monitored at a high salinity for competitive purposes. (C) Cultures of strain JH642 (opuE+) (●), BLOB9 [Δ(opuE::tet)_1_] (▲), and BLOB26 [Δ(opuE::tet)1 × pBLOB15.2 (opuE+ sapB+)] (△) were cultivated in SMM with various salinities for 16 h at 37°C, and the optical densities of the cultures were then determined. The data shown represent a typical set of growth experiments.
Fig 3
Detection of proline released from B. subtilis cells by bioassays. (A) Five-microliter aliquots of JH642 (opuE+), BLOB9 [Δ(opuE::tet)1_], and BLOB26 [Δ(opuE::tet)1 × pBLOB15.2 (opuE+ sapB+)] were spotted onto a lawn of an E. coli lacIZYA+ proline auxotrophic derivative (proC46::Tn_5) of strain MG1655 seeded in top agar. The MMA plates and top agar contained 0.8 M NaCl to osmotically stress B. subtilis and thereby trigger osmoadaptive proline synthesis; IPTG and X-Gal were included in the plates to induce the lac operon and to visualize β-galactosidase activity of this indicator strain. Cross-feeding of the E. coli Pro− auxotroph by proline released from the B. subtilis strain BLOB9 [Δ(opuE::tet)_1_] is visible as a blue halo around the spotted B. subtilis cells. (B) The E. coli proline auxotrophic strain RC711 (proA23) was plated onto a lawn of MMA plates containing 0.8 M NaCl, and B. subtilis colonies were replica plated onto this cell lawn. Cross-feeding of strain RC711 by proline released by the opuE mutant strain BLOB9 is evident from growth of E. coli cells around individual B. subtilis colonies. (C) Blue halo bioassay with strains JH642 (opuE+), BLOB9 [Δ(opuE::tet)_1_] and the mechanosensitive (msc) channel quadruple mutants SMB80 (opuE+) and TMB105 [Δ(opuE::tet)1_], and the E. coli proC46::Tn_5 proline auxotrophic indicator strain.
Fig 4
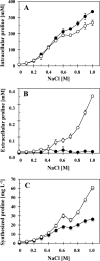
Intracellular and extracellular proline content of high-salinity grown B. subtilis cells. Cultures of the B. subtilis strains JH642 (opuE+) (●) and BLOB9 [Δ(opuE::tet)_1_] (○) were grown in SMM with various salinities. (A) Cells were harvested in mid-exponential growth phase (OD578 of about 1.5), and their proline content was determined by the colorimetric assay described by Bates et al. (2). (B) The same proline assay was also used to determine the proline content of the supernatants of these cultures. (C) The amounts of intracellular and extracellular proline were summed up for culture of strains JH642 and BLOB9 and are given as synthesized proline; we normalized these values according to an OD578 of 1.0. The error bars given are standard deviations of four separately grown B. subtilis cultures (n = 4).
Fig 5
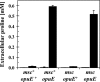
Release of proline into the growth medium is not mediated by MscL- and MscS-type channel proteins. Cells of strains JH642 (msc+ opuE+), BLOB9 (msc+ opuE), SMB80 (msc opuE+), and TMB105 (msc opuE) were grown to mid-exponential growth phase (OD578 of about 2) in either SMM (gray bars) or SMM containing 1 M NaCl (black bars). The proline content of the supernatants was determined by the colorimetric assay described by Bates et al. (2). The error bars given are standard deviations of two separately grown B. subtilis cultures (n = 2). msc mutant strains carry gene disruptions in the mscL, ykuT, yhdY, and yfkC loci, thereby interrupting the single MscL-type channel-forming protein and the three MscS-type channel-forming proteins from B. subtilis (24, 58). opuE mutant strains carry the Δ(opuE::tet)1 allele (56).
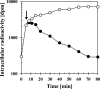
Fig 6
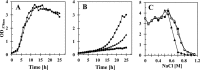
Efflux of [14C]proline from high-salinity-stressed B. subtilis cells in response to an exogenous supply of an excess proline. A culture of the wild-type strain JH642 was grown in SMM with 0.8 M NaCl to mid-exponential growth phase (OD578 of about 1) and
l
-proline that was spiked with 0.63 μM [14C]proline was added to a final concentration of 1 mM. After a 10-min incubation of the cultures (indicated by an arrow), 100 mM proline was added to one culture (●) and 100 mM glycine was added to the second culture (○), and both cell suspensions were then further incubated with vigorous shaking at 37°C. Samples were withdrawn at various time intervals, the cells were collected by filtration onto cellulose filters, and the intracellular [14C]proline content of the cells was determined by scintillation counting. This experiment was repeated three times, and the data given represent a typical result.
Similar articles
- Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B.
von Blohn C, Kempf B, Kappes RM, Bremer E. von Blohn C, et al. Mol Microbiol. 1997 Jul;25(1):175-87. doi: 10.1046/j.1365-2958.1997.4441809.x. Mol Microbiol. 1997. PMID: 11902719 - Osmoregulation of the opuE proline transport gene from Bacillus subtilis: contributions of the sigma A- and sigma B-dependent stress-responsive promoters.
Spiegelhalter F, Bremer E. Spiegelhalter F, et al. Mol Microbiol. 1998 Jul;29(1):285-96. doi: 10.1046/j.1365-2958.1998.00929.x. Mol Microbiol. 1998. PMID: 9701821 - OpuF, a New Bacillus Compatible Solute ABC Transporter with a Substrate-Binding Protein Fused to the Transmembrane Domain.
Teichmann L, Kümmel H, Warmbold B, Bremer E. Teichmann L, et al. Appl Environ Microbiol. 2018 Oct 1;84(20):e01728-18. doi: 10.1128/AEM.01728-18. Print 2018 Oct 15. Appl Environ Microbiol. 2018. PMID: 30097444 Free PMC article. - Guardians in a stressful world: the Opu family of compatible solute transporters from Bacillus subtilis.
Hoffmann T, Bremer E. Hoffmann T, et al. Biol Chem. 2017 Feb 1;398(2):193-214. doi: 10.1515/hsz-2016-0265. Biol Chem. 2017. PMID: 27935846 Review. - Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments.
Kempf B, Bremer E. Kempf B, et al. Arch Microbiol. 1998 Oct;170(5):319-30. doi: 10.1007/s002030050649. Arch Microbiol. 1998. PMID: 9818351 Review.
Cited by
- Contribution of mechanosensitive channels to osmoadaptation and ectoine excretion in Halomonas elongata.
Vandrich J, Pfeiffer F, Alfaro-Espinoza G, Kunte HJ. Vandrich J, et al. Extremophiles. 2020 May;24(3):421-432. doi: 10.1007/s00792-020-01168-y. Epub 2020 Apr 7. Extremophiles. 2020. PMID: 32266565 Free PMC article. - Homeostasis and catabolism of choline and glycine betaine: lessons from Pseudomonas aeruginosa.
Wargo MJ. Wargo MJ. Appl Environ Microbiol. 2013 Apr;79(7):2112-20. doi: 10.1128/AEM.03565-12. Epub 2013 Jan 25. Appl Environ Microbiol. 2013. PMID: 23354714 Free PMC article. Review. - Role of the Extremolytes Ectoine and Hydroxyectoine as Stress Protectants and Nutrients: Genetics, Phylogenomics, Biochemistry, and Structural Analysis.
Czech L, Hermann L, Stöveken N, Richter AA, Höppner A, Smits SHJ, Heider J, Bremer E. Czech L, et al. Genes (Basel). 2018 Mar 22;9(4):177. doi: 10.3390/genes9040177. Genes (Basel). 2018. PMID: 29565833 Free PMC article. Review. - Stress-induced activation of the proline biosynthetic pathway in Bacillus subtilis: a population-wide and single-cell study of the osmotically controlled proHJ promoter.
Morawska LP, Detert Oude Weme RGJ, Frenzel E, Dirkzwager M, Hoffmann T, Bremer E, Kuipers OP. Morawska LP, et al. Microb Biotechnol. 2022 Sep;15(9):2411-2425. doi: 10.1111/1751-7915.14073. Epub 2022 May 20. Microb Biotechnol. 2022. PMID: 35593133 Free PMC article. - Stress responses of the industrial workhorse Bacillus licheniformis to osmotic challenges.
Schroeter R, Hoffmann T, Voigt B, Meyer H, Bleisteiner M, Muntel J, Jürgen B, Albrecht D, Becher D, Lalk M, Evers S, Bongaerts J, Maurer KH, Putzer H, Hecker M, Schweder T, Bremer E. Schroeter R, et al. PLoS One. 2013 Nov 15;8(11):e80956. doi: 10.1371/journal.pone.0080956. eCollection 2013. PLoS One. 2013. PMID: 24348917 Free PMC article.
References
- Alcayaga C, Venegas R, Carrasco A, Wolff D. 1992. Ion channels from the Bacillus subtilis plasma membrane incorporated into planar lipid bilayers. FEBS Lett. 311:246–250 - PubMed
- Bates LS, Waldren RP, Teare ID. 1973. Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
- Boos W, Ehmann U, Bremer E, Middendorf A, Postma P. 1987. Trehalase of Escherichia coli. Mapping and cloning of its structural gene and identification of the enzyme as a periplasmic protein induced under high osmolarity growth conditions. J. Biol. Chem. 262:13212–13218 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases