Standing genetic variation and the evolution of drug resistance in HIV - PubMed (original) (raw)
Standing genetic variation and the evolution of drug resistance in HIV
Pleuni Simone Pennings. PLoS Comput Biol. 2012.
Abstract
Drug resistance remains a major problem for the treatment of HIV. Resistance can occur due to mutations that were present before treatment starts or due to mutations that occur during treatment. The relative importance of these two sources is unknown. Resistance can also be transmitted between patients, but this process is not considered in the current study. We study three different situations in which HIV drug resistance may evolve: starting triple-drug therapy, treatment with a single dose of nevirapine and interruption of treatment. For each of these three cases good data are available from literature, which allows us to estimate the probability that resistance evolves from standing genetic variation. Depending on the treatment we find probabilities of the evolution of drug resistance due to standing genetic variation between 0 and 39%. For patients who start triple-drug combination therapy, we find that drug resistance evolves from standing genetic variation in approximately 6% of the patients. We use a population-dynamic and population-genetic model to understand the observations and to estimate important evolutionary parameters under the assumption that treatment failure is caused by the fixation of a single drug resistance mutation. We find that both the effective population size of the virus before treatment, and the fitness of the resistant mutant during treatment, are key-arameters which determine the probability that resistance evolves from standing genetic variation. Importantly, clinical data indicate that both of these parameters can be manipulated by the kind of treatment that is used.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
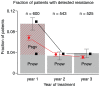
Figure 1. Probability of detecting resistance per year of treatment.
The probability that resistance is detected for the first time in the first, second or third year of treatment, given that it was not detected until then. Grey bars are the estimates from the Margot et al ([36]) dataset, and the number of patients on which the estimates are based are noted at the top of the graph. The red dashed area reflects the inferred probability that resistance mutations from standing genetic variation become established. The black squares are values calculated using equations 2 and 4. The red circles are estimated from 1000 simulations. Parameters as in table 2.
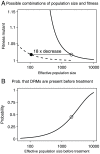
Figure 2. Possible combinations of population size and fitness and the effect of population sizes on the probability that DRMs are present before treatment.
Figure 2a: Continuous line: combinations of population size before treatment ( ) and fitness of mutant virus during therapy (
) and fitness of mutant virus during therapy ( ) that lead to the observed probability that resistance mutations from standing genetic variation become established (
) that lead to the observed probability that resistance mutations from standing genetic variation become established ( ). Dashed line: combinations of population size during treatment (
). Dashed line: combinations of population size during treatment ( ) and fitness of mutant virus during therapy (
) and fitness of mutant virus during therapy ( ) that lead to the observed probability that resistance mutations from standing genetic variation become established (
) that lead to the observed probability that resistance mutations from standing genetic variation become established ( ). Open dot:
). Open dot:  and
and  , closed dot:
, closed dot:  ,
,  . Figure 2b: Probability that a patient has any pre-existing DRMs before the start of therapy for different population sizes, and
. Figure 2b: Probability that a patient has any pre-existing DRMs before the start of therapy for different population sizes, and  . Open dot:
. Open dot:  .
.
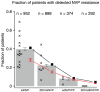
Figure 3. The probability that resistance mutations are detected in women treated for prevention of mother-to-child transmission.
The probability that resistance mutations are detected 6 to 8 weeks after treatment with single dose nevirapine. Black crosses are data from single studies, grey bars with estimated standard error are percentages for all studies combined (the number of patients that were used to calculate this percentage is indicated at the top of the graph). Red circles with standard error are results from 1000 simulations and the black squares are analytical predictions. Parameter values as in table 2.
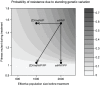
Figure 4. Probability of the establishment of DRMs as a function of effective population size and the fitness of the resistant mutant during treatment.
The predicted probability of the establishment of drug resistance mutations from standing genetic variation depending on the effective population size and the fitness of the resistant mutant during therapy. Grey scales indicate the probability of the evolution of drug resistance due to standing genetic variation. Dots indicate estimated parameter combinations for treatment with just sdNVP, with ZDV monotherapy followed by sdNVP (ZDV/sdNVP), with sdNVP followed by two additional drugs postpartum (sdNVP/PP) and with ZDV monotherapy followed by sdNVP and two additional drugs postpartum ZDV/sdNVP/PP.
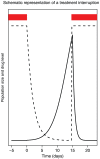
Figure 5. Drug level and population size during and after a treatment interruption.
Drug level (dashed line) and viral population size (solid line) during and after a treatment interruption. Red bars indicate when drugs are taken.
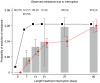
Figure 6. The relationship between the length of a treatment interruption and the probability that DRMs become established.
Estimated probability that resistance mutations become established due to a single treatment interruption. Grey bars are data from seven clinical trials,  estimated standard error (see supplementary table S3 in text S2). The number of patients (and the number of interruptions per patient) are noted at the top of the graph. The red circles are estimated from 1000 simulations,
estimated standard error (see supplementary table S3 in text S2). The number of patients (and the number of interruptions per patient) are noted at the top of the graph. The red circles are estimated from 1000 simulations,  estimated standard error. The black squares are predictions using the average population size from the simulations and equation 2. Parameters as in table 2.
estimated standard error. The black squares are predictions using the average population size from the simulations and equation 2. Parameters as in table 2.
Comment in
- Insufficient Evidence for Rare Activation of Latent HIV in the Absence of Reservoir-Reducing Interventions.
Hill AL, Rosenbloom DI, Siliciano JD, Siliciano RF. Hill AL, et al. PLoS Pathog. 2016 Aug 25;12(8):e1005679. doi: 10.1371/journal.ppat.1005679. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27560936 Free PMC article. No abstract available.
Similar articles
- Nevirapine resistance viral mutations after repeat use of nevirapine for prevention of perinatal HIV transmission.
Kuhn L, Sinkala M, Kankasa MP, Kasonde P, Thea DM, Aldrovandi GM. Kuhn L, et al. J Acquir Immune Defic Syndr. 2006 Jun;42(2):260-2. doi: 10.1097/01.qai.0000214820.26281.df. J Acquir Immune Defic Syndr. 2006. PMID: 16760802 Free PMC article. No abstract available. - Selection and persistence of viral resistance in HIV-infected children after exposure to single-dose nevirapine.
Martinson NA, Morris L, Gray G, Moodley D, Pillay V, Cohen S, Dhlamini P, Puren A, Bhayroo S, Steyn J, McIntyre JA. Martinson NA, et al. J Acquir Immune Defic Syndr. 2007 Feb 1;44(2):148-53. doi: 10.1097/QAI.0b013e31802b920e. J Acquir Immune Defic Syndr. 2007. PMID: 17117145 - Minor drug-resistant HIV type-1 variants in breast milk and plasma of HIV type-1-infected Ugandan women after nevirapine single-dose prophylaxis.
Pilger D, Hauser A, Kuecherer C, Mugenyi K, Kabasinguzi R, Somogyi S, Harms G, Kunz A. Pilger D, et al. Antivir Ther. 2011;16(1):109-13. doi: 10.3851/IMP1698. Antivir Ther. 2011. PMID: 21311114 - Minority HIV-1 drug-resistant mutations and prevention of mother-to-child transmission: perspectives for resource-limited countries.
Samuel R, Paredes R, Parboosing R, Moodley P, Gordon M. Samuel R, et al. AIDS Rev. 2014 Oct-Dec;16(4):187-98. AIDS Rev. 2014. PMID: 25300623 Review.
Cited by
- Evolutionary History and Strength of Selection Determine the Rate of Antibiotic Resistance Adaptation.
Cisneros-Mayoral S, Graña-Miraglia L, Pérez-Morales D, Peña-Miller R, Fuentes-Hernández A. Cisneros-Mayoral S, et al. Mol Biol Evol. 2022 Sep 1;39(9):msac185. doi: 10.1093/molbev/msac185. Mol Biol Evol. 2022. PMID: 36062982 Free PMC article. - Clinical Determinants of HIV-1B Between-Host Evolution and their Association with Drug Resistance in Pediatric Patients.
Pagán I, Rojas P, Ramos JT, Holguín Á. Pagán I, et al. PLoS One. 2016 Dec 1;11(12):e0167383. doi: 10.1371/journal.pone.0167383. eCollection 2016. PLoS One. 2016. PMID: 27907076 Free PMC article. - The evolutionary cost of homophily: social stratification facilitates stable variant coexistence and increased rates of evolution in host-associated pathogens.
Li S, Gulisija D, Carja O. Li S, et al. bioRxiv [Preprint]. 2024 Jul 17:2024.07.14.603415. doi: 10.1101/2024.07.14.603415. bioRxiv. 2024. PMID: 39071438 Free PMC article. Updated. Preprint. - Evolutionary dynamics of HIV at multiple spatial and temporal scales.
Hill AL, Rosenbloom DI, Nowak MA. Hill AL, et al. J Mol Med (Berl). 2012 May;90(5):543-61. doi: 10.1007/s00109-012-0892-1. Epub 2012 May 3. J Mol Med (Berl). 2012. PMID: 22552382 Free PMC article. Review. - Emergence of resistance to atovaquone-proguanil in malaria parasites: insights from computational modeling and clinical case reports.
Cottrell G, Musset L, Hubert V, Le Bras J, Clain J; Atovaquone-Proguanil Treatment Failure Study Group. Cottrell G, et al. Antimicrob Agents Chemother. 2014 Aug;58(8):4504-14. doi: 10.1128/AAC.02550-13. Epub 2014 May 27. Antimicrob Agents Chemother. 2014. PMID: 24867967 Free PMC article.
References
- Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. - PubMed
- Harrigan PR, Whaley M, Montaner JS. Rate of HIV-1 RNA rebound upon stopping antiretroviral therapy. AIDS. 1999;13:F59–F62. - PubMed
- Cozzi-Lepri A, Dunn D, Pillay D, Sabin CA, Fearnhill E, et al. Long-term probability of detecting drug-resistant hiv in treatment-naive patients initiating combination antiretroviral therapy. Clin Infect Dis. 2010;50:1275–1285. - PubMed
- Johnson V, Brun-Vezinet F, Clotet B, Guenthard H, Kuritzkes D, et al. Update of the drug resistance mutations in HIV-1: December 2010. Top HIV Med. 2010;18:156–63. - PubMed
- Mocroft A, Vella S, Benfield TL, Chiesi A, Miller V, et al. Changing patterns of mortality across europe in patients infected with hiv-1. Lancet. 1998;352:1725–1730. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical