Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function - PubMed (original) (raw)
Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function
Reyes Babiano et al. Mol Cell Biol. 2012 Aug.
Abstract
Ribosomal proteins play important roles in ribosome biogenesis and function. Here, we study the evolutionarily conserved L26 in Saccharomyces cerevisiae, which assembles into pre-60S ribosomal particles in the nucle(ol)us. Yeast L26 is one of the many ribosomal proteins encoded by two functional genes. We have disrupted both genes; surprisingly, the growth of the resulting rpl26 null mutant is apparently identical to that of the isogenic wild-type strain. The absence of L26 minimally alters 60S ribosomal subunit biogenesis. Polysome analysis revealed the appearance of half-mers. Analysis of pre-rRNA processing indicated that L26 is mainly required to optimize 27S pre-rRNA maturation, without which the release of pre-60S particles from the nucle(ol)us is partially impaired. Ribosomes lacking L26 exhibit differential reactivity to dimethylsulfate in domain I of 25S/5.8S rRNAs but apparently are able to support translation in vivo with wild-type accuracy. The bacterial homologue of yeast L26, L24, is a primary rRNA binding protein required for 50S ribosomal subunit assembly in vitro and in vivo. Our results underscore potential differences between prokaryotic and eukaryotic ribosome assembly. We discuss the reasons why yeast L26 plays such an apparently nonessential role in the cell.
Figures
Fig 1
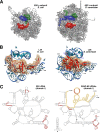
Yeast L26 is the structural homologue of bacterial L24. (A) Localization of L23 (green), L24 (red), and L29 (blue) in the three-dimensional structure of the E. coli 50S subunit and of their respective homologues, L25 (green), L26 (red), and L35 (blue), in the three-dimensional structure of the S. cerevisiae 60S subunit. The cartoons were generated with the UCSF Chimera program, using the atomic model for the crystal structure of the E. coli 70S ribosome (PDB file 3OFC [25]) and the yeast 80S ribosome (PDB files 3U5D and 3U5E [7]). (B) Close-up view of the position of E. coli L24 (left) and yeast L26 (right) in the context of their binding sites in the respective large subunits. Only rRNA residues situated at or closer than 12 Å from L24 or L26 are shown (bases and phosphate backbone). Base numbering follows the 23S rRNA sequence deposited for the E. coli 50S r-subunit (PDB file 3OFC [25]) and the 25S and 5.8S rRNA sequences deposited for the yeast 60S r-subunit (PDB file 3U5D [7]), respectively. Selected residues are labeled in black (23S and 25S rRNAs) or in blue (5.8S rRNA). (C) Secondary structure of domains I from E. coli 23S rRNA and S. cerevisiae 25S/5.8S rRNAs. The structures were taken from The Comparative RNA Web Site (
http://www.rna.icmb.utexas.edu/
) (11). Yeast 5.8S rRNA sequence is highlighted in yellow. Red circles indicate rRNA residues situated closer than 5 Å from E. coli L24 or yeast L26 proteins.
Fig 2
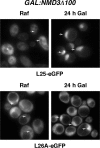
L26 assembles within the nucle(ol)us. Localization of L25-eGFP and L26A-eGFP upon induction of an NMD3 dominant-negative allele; rpl26 null cells expressing L25-eGFP or L26A-eGFP were transformed with the pRS316-GAL-NMD3Δ100 plasmid, and transformants were grown in the presence of raffinose (SRaf-Leu-Ura). Galactose was then added to fully induce the Nmd3Δ100 protein. The GFP signal was inspected by fluorescence microscopy after 24 h. Arrows point to nuclear fluorescence.
Fig 3
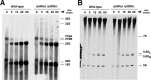
L26-GFP associates with pre-60S ribosomal particles. GFP-tagged L26 was affinity purified with GFP-Trap_A beads from total cellular extracts of rpl26 null cells expressing L26B-eGFP. Wild-type cells were used as an untagged L26 control. RNA was extracted from the pellets obtained after purification (lanes IP) or from an amount of total extracts corresponding to 1/100 of that used for purification (lanes T) and was subjected to Northern analysis of pre- and mature rRNAs. Probes (in parentheses) are described in Fig. S1A and Table S3 in the supplemental material. Signal intensity was measured by phosphorimager scanning; values (below each IP lane) refer to the percentage of each RNA recovered after purification.
Fig 4
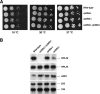
L26 is not essential for growth. (A) The strains BY4741 (wild type), RBY272, a deletant of RPL26A (rpl26aΔ), RBY274, a deletant of RPL26B (rpl26bΔ), and RBY276, an rpl26 null strain (rpl26aΔ rpl26bΔ), were grown in liquid YPD and diluted to an OD600 of 0.05. Serial dilutions were spotted onto YPD plates. Plates were incubated at 30 and 37°C for 3 days or at 16°C for 6 days. (B) Total RNA was extracted from cell extract of the strains, separated by gel electrophoresis, transferred to a nylon membrane, and subjected to Northern analysis. The same filter was consecutively hybridized with α-32P-DNA probes specific for RPL26, RPL35, and ADH1 mRNAs. Mature 25S and 18S rRNAs, which were used as markers to check equal loading, were probed with γ-32P-labeled oligonucleotides (see Fig. S1A and Table S3 in the supplemental material).
Fig 5
Absence of L26 results in a very slight deficit of 60S r-subunits. Polysome profiles are shown for the wild-type and the L26-deficient strains described in Fig. 4. Cells were grown in YPD at 30°C and harvested at an OD600 of around 0.8. Total extracts were prepared, and 10 _A_260 units of each one were resolved on 7 to 50% sucrose gradients. The _A_254 was continuously monitored. Sedimentation is from left to right. The peaks of free 40S and 60S r-subunits, 80S free couples/monosomes, and polysomes are indicated. Half-mer polysomes are labeled by arrows.
Fig 6
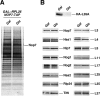
Absence of L26 slightly delays processing of 27SB pre-rRNAs. Wild-type and rpl26 null strains were transformed with the CEN URA3 YCplac33 plasmid and then grown at 30°C in SD-Ura to an OD600 of around 0.8. Cells were pulse-labeled for 2 min with [5,6-3H]uracil and then chased for 5, 15, 30, and 60 min with an excess of unlabeled uracil. Total RNA was extracted, and samples (20,000 cpm per sample) were loaded and separated on a 1.2% agarose–6% formaldehyde gel (A) or a 7% polyacrylamide–8 M urea gel (B), transferred to nylon membranes, and visualized by fluorography. The positions of the different pre-rRNAs and mature rRNAs are indicated.
Fig 7
Absence of L26 slightly alters pre-rRNA processing. The wild-type and L26-deficient strains described in the legend to Fig. 4 were grown in YPD at 30°C and harvested at an OD600 of around 0.8. Total RNA was extracted and subjected to Northern hybridization or primer extension. Probes (in parentheses) are described in Fig. S1A and Table S3 in the supplemental material. (A) Northern analysis of high-molecular-mass pre- and mature rRNAs. (B) Northern analysis of low-molecular-mass pre- and mature rRNAs. (C) Primer extension analysis of 27S pre-rRNAs. Probe f within ITS2 was used.
Fig 8
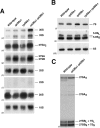
Depletion of L26 does not significantly affect composition of pre-60S ribosomal particles. (A) TAP-tagged Nop7 was used to purify nuclear pre-60S particles from a conditional GAL::RPL26 strain (JWY9634; GAL::HA-RPL26A rpl26bΔ NOP7-TAP) grown in YPGal (Gal) or shifted to YPD (Glc). Copurifying proteins were separated by SDS-PAGE and stained with silver. (B) Western blotting was used to specifically assay the presence of selected ribosome assembly factors and ribosomal proteins in Nop7-TAP-containing preribosomal particles before and after depletion of L26.
Fig 9
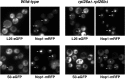
Absence of L26 leads to some nuclear retention of the 60S r-subunit reporter L25-eGFP. Strains BY4741 (wild type) and RBY276 (rpl26aΔ rpl26bΔ) were transformed with plasmids that expressed Nop1-mRFP and either L25-eGFP or S3-eGFP from their cognate promoters. Cells were grown at 30°C in SD-Ura. The subcellular localization of the GFP-tagged ribosomal proteins and the Nop1-mRFP nucleolar marker were analyzed by fluorescence microscopy. Arrows point to nucleolar fluorescence.
Fig 10
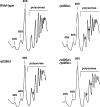
Absence of L26 induces changes in the structure of 25S/5.8S rRNA domain I. (A) In vivo DMS chemical probing experiment of the rRNA structure around the binding site of L26. Wild-type and rpl26 null cells were treated with DMS. Total RNA was extracted from unmodified (−DMS) and modified (+DMS) yeast cells and analyzed by primer extension using the 5.8S_3′END primer (see Table S3 in the supplemental material). U, A, G, and C represent dideoxy sequencing lanes done with the same primer. Stop controls indicate the efficiency of quenching reactions after DMS treatment. Black and green dots denote nucleotides with stronger and weaker reactivity to DMS in the rpl26 null strain, respectively. (B) Secondary structure of yeast 25S/5.8S rRNA domain I (the legend to Fig. 1 provides further details). Red circles indicate nucleotides situated closer than 5 Å from the yeast L26 protein. Nucleotides with stronger and weaker reactivity to DMS in the rpl26 null strain are labeled with black or green arrowheads, respectively (see Fig. S7 in the supplemental material for full DMS probing gels). (C) Close-up view of the three-dimensional arrangement of the yeast 25S/5.8S rRNA domain I, including the ribosomal proteins L17, L35, and L37 (red); L26 (red; the surface is also shown); and L39 (yellow). Only the base side chains of the nucleotides undergoing changes in the DMS reactivity are highlighted and labeled in black or green, as described above. Note that the structure is clipped to simplify the figure.
Similar articles
- Role of the yeast ribosomal protein L16 in ribosome biogenesis.
Espinar-Marchena FJ, Fernández-Fernández J, Rodríguez-Galán O, Fernández-Pevida A, Babiano R, de la Cruz J. Espinar-Marchena FJ, et al. FEBS J. 2016 Aug;283(16):2968-85. doi: 10.1111/febs.13797. Epub 2016 Jul 15. FEBS J. 2016. PMID: 27374275 - Yeast ribosomal protein L40 assembles late into precursor 60 S ribosomes and is required for their cytoplasmic maturation.
Fernández-Pevida A, Rodríguez-Galán O, Díaz-Quintana A, Kressler D, de la Cruz J. Fernández-Pevida A, et al. J Biol Chem. 2012 Nov 2;287(45):38390-407. doi: 10.1074/jbc.M112.400564. Epub 2012 Sep 20. J Biol Chem. 2012. PMID: 22995916 Free PMC article. - The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits.
de la Cruz J, Sanz-Martínez E, Remacha M. de la Cruz J, et al. Nucleic Acids Res. 2005 Oct 12;33(18):5728-39. doi: 10.1093/nar/gki887. Print 2005. Nucleic Acids Res. 2005. PMID: 16221974 Free PMC article. - Ribosome Biogenesis in Plants: From Functional 45S Ribosomal DNA Organization to Ribosome Assembly Factors.
Sáez-Vásquez J, Delseny M. Sáez-Vásquez J, et al. Plant Cell. 2019 Sep;31(9):1945-1967. doi: 10.1105/tpc.18.00874. Epub 2019 Jun 25. Plant Cell. 2019. PMID: 31239391 Free PMC article. Review. - Ribosome biogenesis in the yeast Saccharomyces cerevisiae.
Woolford JL Jr, Baserga SJ. Woolford JL Jr, et al. Genetics. 2013 Nov;195(3):643-81. doi: 10.1534/genetics.113.153197. Genetics. 2013. PMID: 24190922 Free PMC article. Review.
Cited by
- Rpl22 is required for IME1 mRNA translation and meiotic induction in S. cerevisiae.
Kim SJ, Strich R. Kim SJ, et al. Cell Div. 2016 Jul 29;11:10. doi: 10.1186/s13008-016-0024-3. eCollection 2016. Cell Div. 2016. PMID: 27478489 Free PMC article. - Pol5 is an essential ribosome biogenesis factor required for 60S ribosomal subunit maturation in Saccharomyces cerevisiae.
Ramos-Sáenz A, González-Álvarez D, Rodríguez-Galán O, Rodríguez-Gil A, Gaspar SG, Villalobo E, Dosil M, de la Cruz J. Ramos-Sáenz A, et al. RNA. 2019 Nov;25(11):1561-1575. doi: 10.1261/rna.072116.119. Epub 2019 Aug 14. RNA. 2019. PMID: 31413149 Free PMC article. - The RNA helicase Dbp7 promotes domain V/VI compaction and stabilization of inter-domain interactions during early 60S assembly.
Aquino GRR, Hackert P, Krogh N, Pan KT, Jaafar M, Henras AK, Nielsen H, Urlaub H, Bohnsack KE, Bohnsack MT. Aquino GRR, et al. Nat Commun. 2021 Oct 22;12(1):6152. doi: 10.1038/s41467-021-26208-9. Nat Commun. 2021. PMID: 34686661 Free PMC article. - Ribosomal protein eL39 is important for maturation of the nascent polypeptide exit tunnel and proper protein folding during translation.
Micic J, Rodríguez-Galán O, Babiano R, Fitzgerald F, Fernández-Fernández J, Zhang Y, Gao N, Woolford JL, de la Cruz J. Micic J, et al. Nucleic Acids Res. 2022 Jun 24;50(11):6453-6473. doi: 10.1093/nar/gkac366. Nucleic Acids Res. 2022. PMID: 35639884 Free PMC article. - A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains.
Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, Lin L, Woolford JL Jr. Gamalinda M, et al. Genes Dev. 2014 Jan 15;28(2):198-210. doi: 10.1101/gad.228825.113. Genes Dev. 2014. PMID: 24449272 Free PMC article.
References
- Albanèse V, Yam AY, Baughman J, Parnot C, Frydman J. 2006. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124:75–88. - PubMed
- Ausubel FM, et al. 1994. Saccharomyces cerevisiae, p 13.0.1–13.14.17 Current protocols in molecular biology, vol 2 John Wiley & Sons, Inc., New York, NY
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases